Abstract
The Indian false vampire (Megaderma lyra), known as the greater false vampire bat, the Indian false vampire bat, and the greater false-vampire, is typical echolocation mammals. It has been listed in the IUCN Red List of threatened species and included in the Red Book of Endangered Animals in China. Herein, we described 17,055 bp of M. lyra mtDNA that includes 13 protein-coding genes (PGCs), two rRNA genes (12S rRNA and 16S rRNA), 22 transfer RNA (tRNA) genes, and one control region (D-loop). The complete mitochondrial genome sequence will provide new molecular biology information to further understand the genetic diversity of the M. lyra and to protect this population.
The Indian false vampire (Megaderma lyra) belongs to the family Megadermatidae that is widespread throughout South Asia and Southeast Asia (Csorba et al. Citation2008). It is a typical echolocation mammal, and can hunt using both vision and passively listening for its prey, and has also been observed catching prey in complete darkness without echolocation (Reilly Citation2010, Mohd-Azlan et al. Citation2016). However, molecular studies about M. lyra are limited, what’s more, no mitochondrial genome of M. lyra is available until now. Here, we assembled and characterized the complete mitochondrial genome of M. lyra.
The total genomic DNA of M. lyra was isolated from US036, and extracted from the blood of an adult M. lyra, then sequenced with the Genome-wide short-read sequencing (Illumina HisSeq 200, 500 bp insert size, 2 × 90 bp pair-end) (Zhang et al. Citation2014), the information of sample of M. lyra was stored in NCBI (Accession No. SAMN02212695) and the SRA Accession Number is SRS454316. The sequencing reads were trimmed using Trimmomatic v0.36 (Bolger et al. Citation2014), and assembled with NOVOPlasty v2.6.3 software (Dierckxsens et al. Citation2017), then annotated and generated a physical map by MitoFish 3.30 (http://mitofish.aori.u-tokyo.ac.jp/) (Iwasaki et al. Citation2013).
The complete mitochondrial genome of M. lyra is a double-stranded, circular DNA 17,055 bp in total length (GenBank Accession No. MG586969), and includes 13 protein-coding genes, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and one control region (D-loop). The overall nucleotide composition was 30.15% A, 28.32% T, 15.37% C, and 26.13% G, respectively, and the percentage of G + C content was 41.51%. Twelve of the PCGs use complete (TAA) or incomplete (T––) stop codon. The 12S rRNA and 16S rRNA genes are 968 and 1564 bp, respectively. The lengths of 22 tRNA genes range from 59 bp (tRNA-Ser) to 75 bp (tRNA-Leu). The D-loop length is 1151 bp and lies between the ATPase 8 and tRNA-Phe genes.
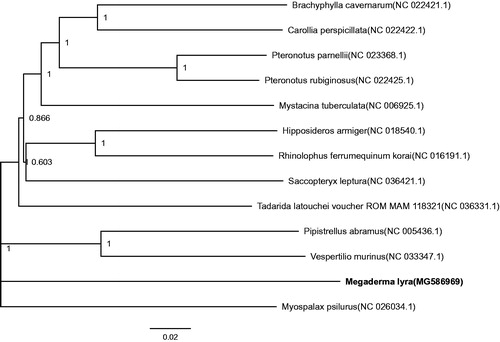
The Phylogenetic analysis of 13 mitochondrial genomes using MEGA 7 (Kumar et al. Citation2016) shows that M. lyra and M. psilurus are the most closely related species (). The mitogenome of M. lyra would contribute to the understanding of the phylogeny and evolution of Rodentia.
Figure 1. Phylogenetic tree constructed with M. lyra and 12 other species mitogenomes and this species belong to Microchiroptera. It was constructed based on the alignment of maximum-likelihood method within the MEGA 7. The bootstrap support values are generated using 1000 replications. GenBank sequences are listed, followed by species names.
Disclosure statement
The authors report no conflicts of interest, and are alone responsible for the content and writing of the paper.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 30:2114.
- Csorba G, Bates P, Molur S, Srinivasulu C. 2008. Megaderma lyra. The IUCN Red List of Threatened Species 2008: e.T12938A3399533. Available from: http://dx.doi.org/10.2305/IUCN.UK.20
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Research. 45:e18.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M. 2013. MitoFish and mitoannotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30: 2531–2540.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionarygenetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Mcginley M. 2012. IUCN Red List of Threatened Species.
- Mohd-Azlan J, Said A, Kheng SL, Tisen OB. 2016. The distribution of Buceros rhinoceros and awareness of its conservation status. In: Das I, Tuen AA, editors. Naturalists, explorers and field scientists in South-East Asia and Australasia. Cham: Springer International Publishing; p. 215–223.
- Reilly SE. 2010. Breeding the rhinoceros hornbill buceros rhinoceros at the Audubon Park and Zoological Garden. International Zoo Yearbook. 27:263–269.
- Zhang G, Cai L, Li Q, Bo L, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 346:1311.
