Abstract
We sequenced the complete mitochondrial genome of Emberiza leucocephala (Pine Bunting; 16,754 bp in length) and E. elegans (Yellow-throated Bunting; 16,780 bp in length). Gene order and orientation are consistent with those of most avian mitochondrial genomes. The result of the phylogenetic analyses suggested that the classification of Melophus and Latoucheornis was monotypic genera, which suggest they should be placed within Emberiza.
We sequenced the complete mitochondrial genome of Emberiza leucocephala and E. elegans that come from Wanyuan, Sichuan province in China (N 32°03′27.72″; E 108°03′45.52″). The specimen was preserved in Key Laboratory of Sichuan Institute for Protecting Endangered Birds in the Southwest Mountains, College of Life sciences, Leshan Normal University, China. Whole muscle tissue preserved in 100% ethanol at −20 °C.
The complete mitogenomes of E. elegans and E. leucocephala is 16,780 bp and 16,754 bp in length, respectively. Base content for E. elegans is as follows: A = 29.33%, T = 23.21%, C = 32.75%, and G = 14.71%. Base content for E. leucocephala is as follows: A = 29.57%, T = 22.96%, C = 32.88%, and G = 14.60%. Both genomes consisted of 13 PCGs, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and a non-coding control region. Gene order and orientation are consistent with those of most avian mitochondrial genomes (Pereira Citation2000; Liu et al. Citation2016).
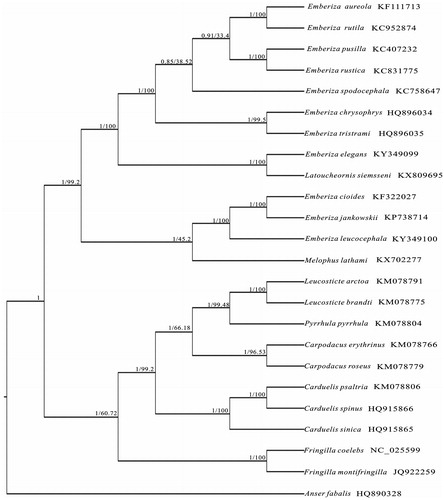
Phylogenetic relationships were reconstructed based on Bayesian inference (BI) and maximum parsimony (MP) method that revealed the same topological phylogenies tree based on the complete mitogenomes of the species (). The results of the phylogenetic analyses suggested that the classification of Melophus and Latoucheornis was monotypic genera, which suggest they should be placed within Emberiza.
Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Liu F, Bao X, Fan Y, Li J, Yao X. 2016. Sequencing and analysis of the complete mitochondrial genome of Brown Shrike, Lanius cristatus (Passeriformes, Laniidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27:3544–3546.
- Pereira SL. 2000. Mitochondrial genome organization and vertebrate phylogenetics. Genet Mol Biol. 23:745–752.