Abstract
The complete mitochondrial genome sequence of the hybrid of Hoplobatrachus chinensis (♀) × H. rugulosus (♂) was obtained in this study. The circular mitochondrial genome was 20,282 bp in length (including extra ND5 genes). Compared with the complete mitogenome of the parents, the results indicated that the mitochondria of the hybrid tiger frog was consistent with a maternal inheritance. Phylogenetic analyses using concatenated nucleotide sequences of the 11 protein-coding genes with two different methods (maximum likelihood and MrBayes analysis) both highly supported a close relationship of the hybrid frogs with the Chinese tiger frog (=H. chinensis).
The numbers of Chinese tiger frogs, Hoplobatrachus chinensis, are now listed in Appendix II of CITES in China. Although many Chinese researchers think that the Chinese tiger frog is correctly designated as H. chinensis, some researchers still believe it to be a synonym of H. rugulosus (Frost Citation2016). Alam et al. (Citation2008), Pansook et al. (Citation2012), and Yu et al. (Citation2015) found that cryptic species existed in the Thailand tiger frog. To evaluate the viable hybrid, we sequenced the complete mitogenome sequence of the H. chinensis (♀) × H. rugulosus (♂) hybrid. The female of H. chinensis was from Jinhua (29°21′39″, 119°57′58″), Zhejiang, China whereas the male H. rugulosus was from the Huwawa farm of Jinhua, Zhejiang, China, originally imported from Thailand. All the samples were stored at –70 °C in the college of Chemistry and Life Science, Zhejiang Normal University. Total genomic DNA stored in the Lab Yu was extracted from the tail of a hybrid tadpole using a standard proteinase K/SDS digest extraction method followed by phenol–chloroform isolation and ethanol precipitation (Sambrook Citation2001). We amplified the entire mitogenome of the hybrid by normal PCR and long-and-accurate PCR (LA-PCR) methods according to Yu et al. (Citation2012, Citation2015). Sequences were checked and assembled using SeqMan (Lasergene version 5.0) (Burland Citation2000). The genomic sequence has been deposited in GenBank with an accession number MG770308.
The complete mitochondrial genome of the hybrid of H. chinensis (♀) × H. rugulosus (♂) was 20,282 bp in length and included an extra copy of ND5 and an extra copy of tRNAMet genes. The two control regions had identical nucleotide sequences, except for an extra TT in 5′-CR2.
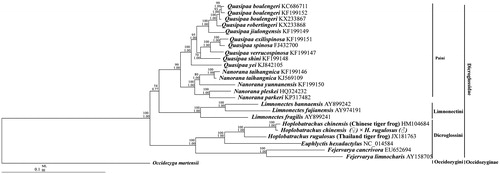
To confirm the phylogenetic relationships between the hybrid of H. chinensis (♀) × H. rugulosus (♂) and other tiger frogs, phylogenetic analyses in MrBayes (BI) and maximum likelihood (ML) analyses were performed on the concatenated nucleotide dataset of 10 PCGs excluding ND5, ATP8, and ND6 genes. Mitogenomes for 25 species from Dicroglossidae were downloaded from GenBank (Liu et al. Citation2005; Ren et al. Citation2009; Zhang et al. Citation2009; Zhou et al. Citation2009; Alam et al. Citation2010; Chen et al. Citation2011; Yu et al. Citation2012; Shan et al. Citation2014; Yu et al. Citation2015; Chen, Zhai, Zhang, et al. Citation2015; Zhang, et al. Citation2018; Chen, Zhai, Zhu, et al. Citation2015; Jiang et al. Citation2016) and used to analyse the phylogenetic relationship of the hybrid using Occidozyga martensii (Li et al. Citation2014) as the outgroup. The tree topologies excluding the branch length produced by BI and ML analyses were equivalent (). The hybrid is a sister clade of H. rugulosus (HM104684, Chinese tiger frog). The hybrid has a closer relationship and smaller genetic distance from its female parent (Chinese tiger frog) than its male parent (Thailand tiger frog), which can be explained by the maternal inheritance characteristics of the mitochondria.
Figure 1. Phylogenetic relationships within Dicroglossidae based on 10 protein-coding genes using nucleotide datasets. Phylogenetic analyses using nucleotide datasets were carried out for the 25 dicroglossids based on all 10 protein-coding genes from their respective mt genomes. The tree was rooted with Occidozyga martensii as the out-group. Numbers above the nodes are the bootstrap values of ML on top and the posterior probabilities of BI on the bottom.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Alam MS, Igawa T, Khan MMR, Islam MM, Kuramoto M, Matsui M, Kurabayashi A, Sumida M. 2008. Genetic divergence and evolutionary relationships in six species of genera Hoplobatrachus and Euphlyctis (Amphibia: Anura) from Bangladesh and other Asian countries revealed by mitochondrial gene sequences. Mol Phylogenet Evol. 48:515–527.
- Alam MS, Kurabayashi A, Hayashi Y, Sano N, Khan MMR, Fujii T, Sumida M. 2010. Complete mitochondrial genomes and novel gene rearrangements in two dicroglossid frogs, Hoplobatrachus tigerinus and Euphlyctis hexadactylus, from Bangladesh. Genes Genet Syst. 85:219–232.
- Burland TG. 2000. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 132:71–91.
- Chen G, Wang B, Liu J, Xie F, Jiang JP. 2011. Complete mitochondrial genome of Nanorana pleskei (Amphibia: Anura: Dicroglossidae) and evolutionary characteristics of the amphibian mitochondrial genomes. Cur Zool. 57:785–805.
- Chen Z, Zhai X, Zhang J, Chen X. 2015. The complete mitochondrial genome of Feirana taihangnica (Anura: Dicroglossidae). Mitochondrial DNA. 26:485–486.
- Chen Z, Zhai X, Zhu Y, Chen X. 2015. Complete mitochondrial genome of the Ye’s spiny-vented frog Yerana yei (Anura: Dicroglossidae). Mitochondrial DNA. 26:489–490.
- Frost DR. 2016. Amphibian species of the world: an online reference. Version 6.0 [accessed 2016 Dec 1]. New York, USA: American Museum of Natural History. http://research.amnh.org/herpetology/amphibia/index.html.
- Jiang L, Ruan Q, Chen W. 2016. The complete mitochondrial genome sequence of the Xizang Plateau frog, Nanorana parkeri (Anura: Dicroglossidae). Mitochondrial DNA A. 27:3184–3185.
- Li E, Li X, Wu X, Feng G, Zhang M, Shi H, Wang L, Jiang J. 2014. Complete nucleotide sequence and gene rearrangement of the mitochondrial genome of Occidozyga martensii. J Genet. 93:631–641.
- Liu ZQ, Wang YQ, Su B. 2005. The mitochondrial genome organization of the rice frog, Fejervarya limnocharis (Amphibia: Anura): a new gene order in the vertebrate mtDNA. Gene. 346:145–151.
- Pansook A, Khonsue W, Piyapattanakorn S, Pariyanonth P. 2012. Phylogenetic relationships among Hoplobatrachus rugulosus in Thailand as inferred from mitochondrial DNA sequences of the cytochrome-b gene (Amphibia, Anura, Dicroglossidae). Zool Sci. 29:54–59.
- Ren Z, Zhu B, Ma E, Wen J, Tu T, Cao Y, Hasegawa M, Zhong Y. 2009. Complete nucleotide sequence and gene arrangement of the mitochondrial genome of the crab-eating frog Fejervarya cancrivora and evolutionary implications. Gene. 441:148–155.
- Sambrook J. 2001. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press.
- Shan X, Xia Y, Zheng YC, Zou FD, Zeng XM. 2014. The complete mitochondrial genome of Quasipaa boulengeri (Anura: Dicroglossidae). Mitochondrial DNA. 25:83–84.
- Yu D, Zhang J, Li P, Zheng R, Shao C. 2015. Do cryptic species exist in Hoplobatrachus rugulosus? An examination using four nuclear genes, the Cyt b gene and the complete MT genome. PLoS One. 10:e0124825.
- Yu DN, Zhang JY, Zheng RQ, Shao C. 2012. The complete mitochondrial genome of Hoplobatrachus rugulosus (Anura: Dicroglossidae). Mitochondrial DNA. 23:336–337.
- Zhang JF, Nie LW, Wang Y, Hu LL. 2009. The complete mitochondrial genome of the large-headed frog, Limnonectes bannaensis (Amphibia: Anura), and a novel gene organization in the vertebrate mtDNA. Gene. 442:119–127.
- Zhang JY, Zhang LP, Yu DN, Storey KB, Zheng RQ. 2018. Complete mitochondrial genomes of Nanorana taihangnica and N. yunnanensis (Anura: Dicroglossidae) with novel gene arrangements and phylogenetic relationship of Dicroglossidae. BMC Evol Biol. 18:26.
- Zhou Y, Zhang JY, Zheng RQ, Yu BG, Yang G. 2009. Complete nucleotide sequence and gene organization of the mitochondrial genome of Paa spinosa (Anura: Ranoidae). Gene. 447:86–96.
