Abstract
The loggerhead turtle, Caretta caretta (Linnaeus, 1758), is an endangered sea turtle in Colombian Caribbean beach. In the present study was sequenced the complete mitochondrial DNA of three loggerhead turtles using Illumina next-generation sequencing (NGS). The average nucleotide frequency was A: 35% T: 26%, C: 27% and G: 12%. This genome provides knowledge to the study of genetic variations and evolution of mitochondrial genomes of C. caretta. The sequences were deposited at the GenBank database under the accession number MF554690.1, MF579504.1 and MF579505.1.
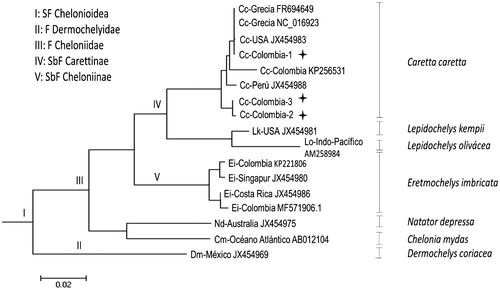
The loggerhead turtle is a marine turtle belonging to the Cheloniidae family, order Testudines. Loggerhead turtle is a species widely distributed in the tropical and Central Atlantic and Indo-Pacific region (Lancheros-Piliego and Fernández Citation2013). Due to the population decrease of this species in the oceans around the world, generated by anthropic actions such as egg consumption, use of shells, commercial fishing and destruction of natural habitats, among others (Lutcavage et al. Citation1997), survival of loggerhead turtles is considered Vulnerable A2b globally (IUCN Citation2017) and Critically Endangered A2cd; D in Colombia (Páez et al. Citation2015). To generate conscience and contribute to its conservation, phylogenetics, genetics of populations and evolution analyses have been carried out using mitochondrial DNA. Blood samples from three individuals of loggerhead turtle were obtained from the CEINER Oceanarium in San Martin de Pajares Island, Cartagena (10°11′N, 75°47′W) following the Dutton methodology (Dutton Citation1996). The blood samples were used for total RNA extraction using RNeasy Mini Kit (Quiagen, Hilden, Germany). For mRNA library preparation, we use a TruSeq RNA Library Prep Kit v2 according to manufacturer’s instructions (Illumina, San Diego, CA, USA). The quality control of generated libraries was done using the 2100 bioanalyzer (Agilent, Santa Clara, CA, USA). RIN values (RNA integrity number) of 7.5 were obtained. The library was paired-end sequenced using Hiseq 2000 Platform (Macrogen, Inc, Korea) (Hernández-Fernández et al. 2017). The quality of cleaned raw reads was verified with the fastQC program (http://www.bioinformatics.bbrc.ac.uk/projects/fastqc/). Using the Local Basic Alignment Search Tool (BLASTn) the contigs of each transcriptome were filtered to establish the complete mitogenome of each individual by aligning the local data obtained from the sequencing against a mitochondrial genome of C. caretta previously reported in GenBank (NCBI) (access number: NC_016923. 1). For each consensus sequence, paired alignments were made with each of the 37 genes encoded by the mtDNA (reported in GenBank) using the BLAST refseq_genomic tool (NCBI). The phylogenetic tree was done using software MEGA 6 (Tamura et al. Citation2013) using the maximum likelihood algorithm (Guindon and Gascuel, Citation2003) (). The three complete mitogenomes of loggerhead turtles were annotated, they comprised 16,633, 16,461 and 16,446 bp, composed for 13 protein-coding genes, 22 tRNA, 2 rRNA and a non-coding control region. These mitogenomes have a typical order of terrestrial, freshwater, and marine turtles (Drosopoulou et al. Citation2012; Duchene et al. Citation2012) and vertebrates in general (Boore, Citation1999). Our analysis of the loggerhead mitogenomes is the first report in the Colombian Caribbean, generating new basic information for studies at a genetic level and contributing to management and conservation suitable plans for this species.
Acknowledgements
We are grateful to CEINER Oceanarium on the island of St. Martin Pajares for their collaboration in obtaining and providing samples of Caretta caretta for the development of this study. Samples were obtained under a research permit that was granted by the Ministry of Environment and Territorial Development (#24 of June 22, 2012) and Contract for Access to Genetic Resources (#64 of April 23, 2013).
Disclosure statement
The authors report no conflict of interest and also state that they are responsible for content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Research. 27:1767–1780.
- Drosopoulou E, Tsiamis G, Mavropoulou M, Vittas S, Katselidis KA, Schofield G, Palaiologou D, Sartsidis T, Bourtzis K, Pantis J, et al. 2012. The complete mitochondrial genome of the loggerhead turtle Caretta caretta (Testudines: Cheloniidae): genome description and phylogenetic considerations. Mitochondrial DNA. 23:1–12.
- Duchene S, Frey A, Alfaro-Núñez A, Dutton PH, Gilbert MT, Morin PA. 2012. Marine turtle mitogenome phylogenetics and evolution. Mol Phylogenet Evol. 65:241–250.
- Dutton P. 1996. Methods for collection and preservation of samples for sea turtle genetic studies, Proceedings of the International Symposium on Sea Turtle Conservation Genetics, NOAA Tech; 1996; Memo, Miami, Florida, p. 17–24 NMFS-SEFSC- 396.
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 52:696–704.
- Hernández-Fernández J, Pinzón A, Mariño-Ramírez L. 2017. De novo transcriptome assembly of loggerhead sea turtle nesting of the Colombian Caribbean. Genomics Data. 13:18–20.
- IUCN. 2017. IUCN Red List of Threatened Species. Versión 2016.4. [cited 2017 Oct 10]. www.iucnredlist.org.
- Lancheros-Piliego D, Fernández JH. 2013. AMDAR y PCR-extra-rápida para la identificación de la tortuga cabezona Caretta caretta (Testudines: Cheloniidae) utilizando el gen mitocondrial citocromo c oxidasa I (COI). Universitas Scientiarum. 18:321.
- Lutcavage M, Plotkin P, Witherington B, Lutz P. 1997. Human impacts on sea turtle survival. En: The biology of sea turtles. Vol. 1. In: Lutz PL and Musick JA, editors. Boca Raton: CRC Press, p. 387–410 (Miscellaneous, Survival).
- Páez V, Ramírez-Gallego C, Barrientos-Muñoz K. 2015. Caretta caretta (Linnaeus, 1758) Pp. 118–121. In: Morales-Betancourt M, Lasso C, Páez VY, Bock B, editors. Libro rojo de reptiles de Colombia (2015). Instituto de Investigación de Recursos Biológicos Alexander von Humboldt (IAvH).Bogotá, D.C., Colombia: Universidad de Antioquia.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.