Abstract
The complete mitochondrial genome (mitogenome) of Pidorus atratus (Lepidoptera: Zygaenoidea: Zygaenidae) is described in this study. The circular molecule is 15,383 bp in length and contains 37 typical mitochondrial genes and one non-coding AT-rich region. All protein-coding genes (PCGs) start with ATN, except for cox1 gene with CGA; 10 of the 13 PCGs harbour the typical stop codon TAA, whereas cox1, cox2, and nad4 end with a single T. Two ribosomal RNA genes (rRNAs) are 1366 bp and 544 bp in length, respectively. The AT-rich region is 658 bp in size and harbours several features characteristic of the lepidopterans, including the motif ATAGA followed by a 20 bp poly-T stretch and a microsatellite-like (TA)9 element. The complete mitogenome data would be useful for further understanding the taxonomy and phylogeny of Zygaenoidea.
Zygaenoidea is one of the superfamilies of lepidopterans, consisting of about 12 families, including the Zygaenidae and Limacodidae (van Nieukerken et al. Citation2011). Due to their unique biological characteristics, Zygaenoidea is of great interest to lepidopterists and evolutionary biologists, with its internal phylogeny and phylogenetic relationships with its closely related lepidopteran groups still standing as a controversial issue (Liu et al. Citation2016). As far as we knew, up to the present, more than 200 complete or near-complete lepidopteran mitogenomes have been determined, however, among these determined mitogenomes, only four are for the Zygaenoidea moths (Tang et al. Citation2014; Liu et al. Citation2016; Peng et al. Citation2017). In this study, we first reported the complete mitogenome of Pidorus atratus (Lepidoptera: Zygaenoidea: Zygaenidae). Adult individuals of P. atratus were collected from Mountain Huangshan, Anhui province, China in August 2017. After morphological identification, the specimens were kept in the laboratory at –80 °C until used for DNA extraction. Total genomic DNA was isolated and purified from a single leg using a DNA extraction kit (Sangon Biotech Co., Ltd, Shanghai, China) according to the manufacturer’s instructions. All the PCRs and PCR product sequencings were performed after Hao et al. (Citation2013) and the resultant reads were assembled and annotated using the BioEdit 7.0 (Hall Citation1999) and MEGA 7.0 (Kumar et al. Citation2016).
The complete mitogenome of P. atratus is a circular molecule of 15,383 bp in size (GenBank accession MG882482), containing typical 37 genes for insects: 13 protein-coding genes (PCGs), 22 tRNAs, two ribosomal RNA genes (rRNAs), and one AT-rich region. The gene order and arrangement are consistent with all other lepidopterans (e.g. Yang et al. Citation2009; Sun et al. Citation2012; Hao et al. Citation2013; Wu et al. Citation2016). The A + T content of the whole mitogenome is 79.8%, which is generally in accordance with other four determined Zygaenoidea mitogenomes. All PCGs use typical ATN as start codon, except the cox1 which uses CGA as its start codon; 10 PCGs use the standard stop codon TAA, while cox1, cox2, and nad4 end with a single T. All tRNAs are folded into the cloverleaf secondary structures except tRNASer (AGN) which lacks the DHU arm. The rrnL (16S rRNA) and rrnS (12S rRNA) genes are 1366 bp and 544 bp in size, respectively. The putative AT-rich region is 658 bp in size, harbouring several structures characteristic of lepidopterans (Hao et al. Citation2013; Shi et al. Citation2015).
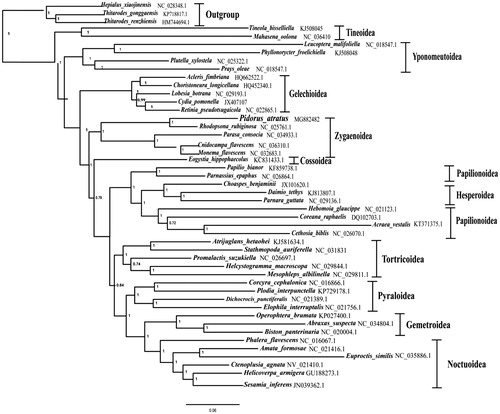
The phylogenetic tree of 44 lepidopteran species including the P. atratus of this study and other four determined Zygaenoidea species, which represent 11 lepidopteran superfamilies were reconstructed based on the concatenated 13 PCG nucleotide sequence data with the Bayesian inference (BI) method, using three Hepialidae species as the outgroups (see for details). The result showed the P. atratus formed a monophyletic group with other four Zygaenoidea species with strong support value, which contradicted with the result of previous study in that the two Zygaenoidea families, Zygaenidae and Limacodidae, constituted a paraphyletic group (Liu et al. Citation2016).
Figure 1. The Bayesian inference (BI) phylogenetic analysis tree of P. atratus and other lepidopterans. Phylogenetic reconstruction was done from a concatenated matrix of 13 protein-coding mitochondrial genes of the mitochondrial genome. The numbers beside the nodes are bootstrap values. Alphanumeric terms indicate the GenBank accession numbers.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this paper.
Additional information
Funding
References
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Hao JS, Sun ME, Shi QH, Sun XY, Shao LL, Yang Q. 2013. Complete mitogenomes of Euploea muciber (Nymphalidae: Danainae) and Libythea celtis (Nymphalidae: Libytheinae) and their phylogenetic implications. ISRN Genomics. 2013:1–14.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu QN, Xin ZZ, Bian DD, Chai XY, Zhou CL, Tang BP. 2016. The first complete mitochondrial genome for the subfamily Limacodidae and implications for the higher phylogeny of Lepidoptera. Sci Rep. 6:35878.
- Peng SY, Zhang Y, Zhang XC, Li Y, Huang ZR, Zhang YF, Zhang X, Ding JH, Geng XX, Li J. 2017. Complete mitochondrial genome of Cnidocampa flavescens (Lepidoptera: Limacodidae). Mitochondrial DNA B. 2:534–535.
- Shi QH, Sun XY, Wang YL, Hao JS, Yang Q. 2015. Morphological characters are compatible with mitogenomic data in resolving the phylogeny of nymphalid butterflies (Lepidoptera: Papilionoidea: Nymphalidae). PLoS One. 10:e0124349.
- Sun QQ, Sun XY, Wang XC, Gai YH, Hu J, Zhu CD, Hao JS. 2012. Complete sequence of the mitochondrial genome of the Japanese buff-tip moth, Phalera flavescens (Lepidoptera: Notodontidae). Genet Mol Res. 11:4213–4225.
- Tang M, Tan MH, Meng GL, Yang SZ, Su X, Liu SL, Song WH, Li YY, Wu Q, Zhang AB, et al. 2014. Multiplex sequencing of pooled mitochondrial genomes: a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res. 42:e166.
- Wu YP, Zhao JL, Su TJ, Luo AR, Zhu CD. 2016. The complete mitochondrial genome of Choristoneura longicellana (Lepidoptera: Tortricidae) and phylogenetic analysis of Lepidoptera. Gene. 591:161–176.
- Yang L, Wei ZJ, Hong GH, Jiang ST, Wen LP. 2009. The complete nucleotide sequence of the mitochondrial genome of Phthonandria atrilineata (Lepidoptera: Geometridae). Mol Biol Rep. 36:1441–1449.
- van Nieukerken EJ, Kaila L, Kitching IJ, Kristensen NP, Lees DC, et al. 2011. TOrder Lepidoptera. In: Zhang ZQ. (Ed.) Animal biodiversity: An outline of higher-level classification and survey of taxonomic richness. Zootaxa. 3148:212–221.
