Abstract
Here, we present the complete mitochondrial genome of the sakura shrimp, Sergia lucens (Crustacea, Decapoda, Sergestidae). The circular genome is 16,087 base pairs in length and contains 13 protein-coding genes, 22 tRNA genes, and two rRNA genes. The nucleotide composition of the S. lucens mitogenome is biased towards A + T (69.9%). The gene arrangement was identical to penaeid shrimps. Maximum likelihood phylogenetic analysis using the nucleotide sequences of 13 protein-coding genes placed S. lucens next to Acetes chinensis, supporting the conventional taxonomic relationship of Sergia and Acetes.
The sakura shrimp Sergia lucens is a small deep-sea crustacean that is primarily distributed to the waters off Japan and Taiwan (Imai et al. Citation2013). It has been an important fishery target in Suruga Bay, Japan. The genus Sergia belongs to the family Sergestidae, a group of pelagic shrimps that boast large biomass in temperate to tropical waters worldwide (Omori Citation1975). Despite the ecological and economic importance of Sergestidae, only one complete mitogenome (Acetes chinensis; GenBank accession no. NC_017600) has been reported. To overcome the deficit of sergestoid genetic information, we sequenced the whole mitogenome of S. lucens, one of the most economically important sergestoid shrimps.
A commercial supplier provided frozen S. lucens specimens that were sampled from a fishing ground in Suruga Bay, Japan. The exact geographic coordinate of the sampling site is unknown because the specimens were caught by trawling. We extracted genomic DNA from a whole shrimp body using the standard phenol–chloroform protocol. The DNA sample of the sequenced specimen is available from the authors. Intact specimens derived from the same sampling batch (MTUF-Ar 00016) are also available in the Marine Science Museum, Tokyo University of Marine Science and Technology. We constructed a paired-end DNA library using Nextera XT library preparation kit (Illumina, San Diego, CA). We sequenced the library with the MiSeq platform (Illumina, San Diego, CA) and obtained 5.8 million paired-end reads with a total of 720 million bases. De novo assembly of read data using CLC Genomics Workbench 6.5.1 (Qiagen, Germany) generated a putative mitogenome sequence, which we verified by Sanger sequencing. We used the MITOS pipeline (Bernt et al. Citation2013) for annotation and manually curated the MITOS output with penaeid mitogenomes. The complete mitogenome of S. lucens is available at DDBJ with accession no. LC368254. The raw Illumina read data can also be found in DDBJ Sequence Read Archive with accession no. DRR121359.
We conducted a maximum likelihood (ML) phylogenetic analysis of S. lucens and other shrimps using the 13 mitochondrial protein-coding genes. We used MAFFT version 7.310 (Katoh and Standley Citation2013) to generate multiple sequence alignments (MSAs) of the 13 protein sequences; the protein MSAs were then used as pgaligncodon 2.0 (Tanabe Citation2011) input to build codon-adjusted nucleotide MSAs. We excluded third codon positions of the protein coding genes and the rRNA sequences from the dataset because they exhibited GC content heterogeneity and may confound the analysis. We used Kakusan 4 (Tanabe Citation2011) to select the partitioning pattern (equal substitution rate among the genes) and the substitution model (GTRGAMMAX) used in RAxML 8.2.9 (Stamatakis Citation2006).
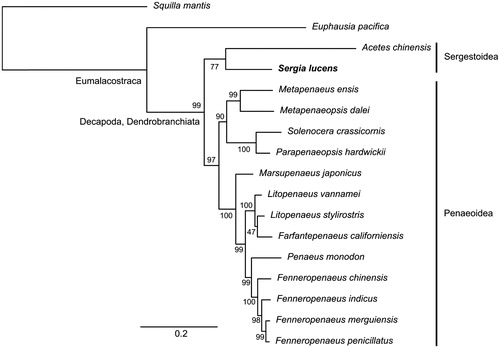
The complete mitogenome of S. lucens was circular and 16,087 bp in length. It contained 13 protein-coding genes, 22 tRNA genes, and two rRNA genes. The nucleotide composition was biased towards A + T rich (69.9%). The gene arrangement of the S. lucens mitogenome was identical to penaeid shrimps. The ML phylogenetic tree clustered S. lucens and A. chinensis in a single branch, albeit with a moderate bootstrapping support (77%), which conforms to the conventional taxonomic relationship of sergestoid shrimps ().
Figure 1. Maximum-likelihood phylogenetic tree of Sergia lucens and other 14 shrimps using two non-decapod crustaceans (Squilla mantis and Euphausia pacifica) as an outgroup. The concatenated nucleotide dataset of 13 mitochondrial protein-coding genes (excluding third codon positions) contained 11,040 sites. The bar below the tree indicates the number of nucleotide substitution per site, and the number beside the branches indicates the bootstrapping support (1000 trials) of each branch. The GenBank accession numbers of the mitogenome sequences used in the analysis are: S. mantis NC_006081; E. pacifica NC_016184; Acetes chinensis NC_017600; Sergia lucens LC368254; Metapenaeus ensis NC_026834; Metapenaeopsis dalei NC_029457; Solenocera crassicornis NC_030280; Parapenaeopsis hardwickii NC_030277; Marsupenaeus japonicus NC_007010; Litopenaeus vannamei NC_009626; L. stylirostris NC_012060; Farfantepenaeus californiensis NC_012738; Penaeus monodon NC_002184; Fenneropenaeus chinensis NC_009679; F. indicus NC_031366; F. merguiensis NC_026884; F. penicillatus NC_026885.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler FP. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Imai H, Hanamura Y, Cheng JH. 2013. Genetic and morphological differentiation in the Sakura shrimp (Sergia lucens) between Japanese and Taiwanese populations. Contrib Zool. 82:123–130.
- Katoh K, Standley MD. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Omori M. 1975. The biology of pelagic shrimps in the ocean. In: Russell SF, Yonge M, editors. Advances in marine biology, vol. 12. London (UK): Academic Press; p. 233–324.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Tanabe AS. 2011. Kakusan4 and Aminosan: two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol Ecol Resour. 11:914–921.
