Abstract
We determined the complete chloroplast DNA sequence of Artemisia hallaisanensis Nakai, an endemic herbal species distributed on Jeju Island, Korea. The chloroplast DNA is 151,015 bp in length and encodes 4 rRNA, 30 tRNA, and 80 protein-coding genes. Phylogenetic analysis and sequence comparison of protein-coding genes with other Artemisa chloroplast DNAs revealed that the chloroplast genome of A. hallaisanensis is closely related to that of A. capillaris. Additionally, a unique 9 bp deletion in ycf1 gene is specific to A. hallaisanensis.
Artemisia is the largest genus in the tribe Anthemideae of the Asteraceae family consisting of more than 500 species that are widely distributed in temperate regions of the northern hemisphere (Oberprieler et al. Citation2009; Vallès et al. Citation2011). The Artemisia species have been well known as medicinal herbs in Asia for their high accumulation of essential oils and terpenoids (Wagner Citation1977). However, taxonomic delimitation of several taxa based on morphological characteristics has been controversial. Molecular marker techniques could solve this issue by providing better criteria for classification of Artemisia species (National Institute of Biological Resources Citation2015).
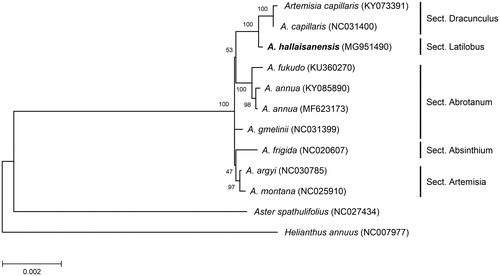
In this study, we determined the complete chloroplast (cp) genome of A. hallaisanensis, an endemic species of Korea showing a very limited distribution on Jeju Island (National Institute of Biological Resources Citation2013). The specimen was collected from Jeju Island (33°21′34″N, 126°30′37.9″E) and deposited in the National Institute of Biological Resources with an accession number NIBR-VP0000538771. A total of 26 million paired-end reads were generated using the Illumina MiSeq platform. A circular DNA was assembled using NOVOPlasty program (version 2.5.9; Dierckxsens et al. Citation2017) with rbcL and rpoC2 genes of A. annua cp DNA (Shen et al. Citation2017) as seed sequences. The assembled cp DNA (Genbank accession MG951490) was 151,015 bp in length with 37.5% overall GC content. The genome structure was highly similar to the reported Artemisia cp DNAs, consisting of two IRs (24,951 bp), one LSC (82,823 bp), and one SSC (18,290 bp). Genome annotation using Geneious program (version 11; Kearse et al. Citation2012) predicted a total of 114 genes (4 rRNA, 30 tRNA, and 80 protein-coding genes), of which 18 genes (4 rRNA, 7 tRNA, and 7 protein-coding genes) were duplicated in two IRs. Twenty genes (6 tRNA and 14 protein-coding genes) contained one or two introns. The maximum likelihood tree constructed using 77 non-redundant protein-coding genes in cp DNAs from 10 Artemisia species and two outgroup species (Aster spathulifolius and Helianthus annuus) indicated that the cp genome of A. hallaisanensis is closely related to that of A. capillaris in section Dracunculus (). Moreover, sequence comparison identified distinct sequence variations in ycf1 gene of A. hallaisanensis and A. capillaris. Two deletions (21 bp and 6 bp, respectively) in ycf1 were evident in both taxa. Interestingly, A. hallaisanensis contains additional 9 bp deletion in ycf1. This unique sequence divergence identified in the cp DNA will serve as a molecular basis for identification of this species as well as phylogenetic study of Artemisia.
Figure 1. Maximum likelihood tree based on the chloroplast protein-coding genes of 12 taxa including A. hallaisanensis. Nucleotide sequences of 77 non-redundant protein-coding genes from publically available Artemisia species as well as two distantly related taxa (Aster spathulifolius and Helianthus annuus) from Asteraceae were aligned and used for maximum likelihood phylogenetic analysis in MEGA7 (Kumar et al. Citation2016) with bootstrap test of 1000 replications. The NCBI accession numbers of chloroplast DNA sequences used in this study are presented in parentheses. The scale bar represents the number of substitutions per site.
Disclosure statement
None of the authors report any conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- National Institute of Biological Resources. 2013. Endemic species of Korea (Plantae). Incheon, Korea: Doo-Hyun Publishing Co.
- National Institute of Biological Resources. 2015. DNA barcode system for Korean indigenous plant species (I). Incheon, Korea: Dong-Jin Publishing Co.
- Oberprieler C, Himmelreich S, Källersjö M, Vallès J, Watson LE, Vogt R, 2009. Anthemideae. In: Funk VA, Susanna A, Steussy TF, et al. editors. Systematics, evolution, and biogeography of compositae. Chapter 38, Madroño: California Botanical Society; p. 631–666.
- Shen X, Wu M, Liao B, Liu Z, Bai R, Xiao S, Li X, Zhang B, Xu J, Chen S. 2017. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules (Basel, Switzerland). 22:1330.
- Vallès J, Garcia S, Hidalgo O, Martín J, Pellicer J, Sanz M, Garnatje T. 2011. Biology, genome evolution, biotechnological issues and research including applied perspectives in Artemisia (Asteraceae). In: Kader J-C, Delseny M, editors. Advances in botanical research. Vol. 60, Chapter 7, London: Academic Press; p. 349–419.
- Wagner H. 1977. Pharmaceutical and economic uses of the compositae. In: Heywood V, Harborne J, Turner B, editors. The biology and chemistry of the compositae. Chapter 14. London: Academic Press; p. 411–433.
