Abstract
The rush sedge caddisfly Phryganea cinerea Walker, 1852 (Phryganeidae, the giant casemakers), is a widespread and adaptable North American caddisfly. Genome skimming by Illumina sequencing permitted the assembly of a complete 15,043 bp circular mitogenome from P. cinerea consisting of 78.2% AT nucleotides, 22 tRNAs, 13 protein-coding genes, 2 rRNAs and a control region in the ancestral insect gene order. Phryganea cinerea COX1 features an atypical CGA start codon and COX1, NAD1, NAD4, and NAD5 exhibit incomplete stop codons completed by the addition of 3′ A residues to the mRNA. Phylogenetic reconstruction reveals a monophyletic Order Trichoptera and Family Phyrganeidae.
The Living Prairie Mitogenomics Consortium is a project to construct a library of arthropod mitogenomes for improved DNA-based species identification and phylogenetics (Living Prairie Mitogenomics Consortium Citation2017; Marcus Citation2018). Mitogenome sequences were annotated by undergraduates in a course inquiry-based learning exercise (Marcus et al. Citation2010). Students who analyzed the data successfully (which were further curated by the instructor) belonged to our consortium.
On 30–31 July 2015, a USDA blacklight trap (Winter Citation2000) was deployed to collect night-flying insects at the Living Prairie Museum (LPM, GPS 49.889607 N, -97.270487 W). Nearby aquatic habitats include Sturgeon Creek (0.57 km) and the Assiniboine River (1.92 km) (Marcus Citation2018). One adult specimen of the rush sedge caddisfly Phryganea cinerea Walker, 1852 (Insecta: Trichoptera: Phryganeidae, the giant casemakers, project specimen number 2015.07.30.014) was trapped, pinned and deposited in the Wallis Roughley Museum of Entomology at the University of Manitoba (voucher JBWM0360830).
Phryganea cinerea larvae are common in North American marshes and littoral zones of lakes (Williams and Penak Citation1980), but are also found at depths of up to 100 m (Selgeby Citation1974) and in sluggish streams (Hilsenhoff Citation1970). Larvae preferentially make cases from shredded vegetation, but will use many other materials (Neave Citation1933; Williams and Penak Citation1980). To lay eggs, adult female P. cinerea will fly up to 10 m in the air and dive-bomb the water surface, sending up small splashes, mimicking rain (LaFontaine Citation1981). Presented here is the first complete New World mitogenome for family Phryganeidae from P. cinerea.
DNA was prepared (McCullagh and Marcus Citation2015) and sequenced by Illumina MiSeq (San Diego, CA) (Peters and Marcus Citation2017). The mitogenome of P. cinerea (Genbank MG980616) was assembled by Geneious 10.1.2 from 6,093,858 paired 75 bp reads using a Eubasilissa regina (Trichoptera: Phryganeidae) reference mitogenome (NC023374) (Wang et al. Citation2014). Annotation was in reference to E. regina and Anabolia bimaculata (Trichoptera: Limnephilidae, MF680449) mitogenomes (Peirson and Marcus Citation2017). The T. tardus nuclear rRNA repeat (Genbank MG986214) was also assembled and annotated using A. bimaculata (MF680448) and Triaenodes tardus (Trichoptera: Leptoceridae, MG201853) (Lalonde and Marcus Citation2017) reference sequences.
The P. cinerea circular 15,043 bp mitogenome assembly was composed of 19,655 paired reads with nucleotide composition: 39.2% A, 14.0% C, 7.7% G, and 39.0% T. Gene composition and order in P. cinerea is identical to most other trichopteran mitogenomes (Marcus Citation2018). Phryganea cinerea COX1 begins with an aberrant start codon (CGA) that is typical of insects (Liao et al. Citation2010). The mitogenome contains four protein-coding genes (COX1, NAD1, NAD4, NAD5) with single-nucleotide (T) stop codons completed by post-transcriptional addition of 3′ A residues. The tRNAs, rRNAs, and control region are typical for Trichoptera (Lalonde and Marcus Citation2017; Peirson and Marcus Citation2017).
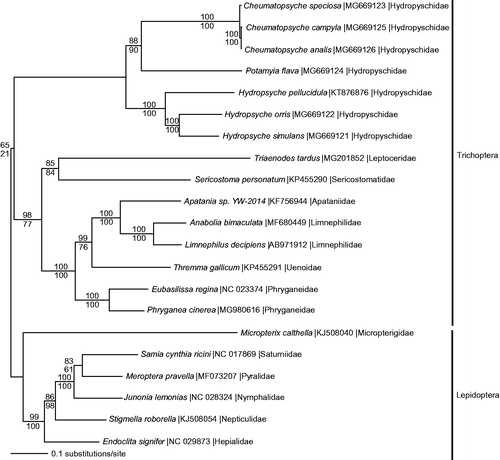
The mitogenomes from P. cinerea, 14 other Trichoptera, and 6 species from sister order Lepidoptera were aligned in CLUSTAL Omega (Sievers et al. Citation2011) and analyzed by maximum likelihood (ML) and parsimony in PAUP* 4.0b8/4.0d78 (Swofford Citation2002) (). Phylogenetic analysis reveals a monophyletic Trichoptera and places P. cinerea as sister to E. regina in monophyletic family Phryganeidae.
Figure 1. Maximum likelihood phylogeny of superorder Amphiesmenoptera (GTR + I + G model, I = 0.1730, G = 0.9090, likelihood score 186552.81293) included complete mitochondrial genome sequences from Phryganea cinerea, 14 other Trichoptera species, and 6 representatives from sister clade Lepidoptera based on 1 million random addition heuristic search replicates (with tree bisection and reconnection). One million maximum parsimony heuristic search replicates also produced a single tree (40,342 steps) with a topology identical to the ML tree. Maximum likelihood bootstrap values are above nodes and maximum parsimony bootstrap values are below nodes (each from 1 million random fast addition search replicates).
Acknowledgements
We thank Sarah Semmler and Kyle Lucyk for permitting and encouraging our work at the Living Prairie Museum. We thank Melissa Peters for help with fieldwork and Aleksandar Ilik and Debbie Tsuyuki (Children’s Hospital Research Institute of Manitoba Next Generation Sequencing Platform) for assistance with library preparation and sequencing.
Disclosure statement
The authors report no conflicts of interest, and are solely responsible for this paper.
Additional information
Funding
References
- Hilsenhoff WL. 1970. Key to genera of Wisconsin Plecoptera (stonefly) nymphs, Ephemeroptera (mayfly) nymphs, and Trichopera (caddisfly) larvae. Wis Dep Nat Resour Res Rep. 67:1–68.
- LaFontaine G. 1981. Caddisflies. New York (NY): Winchester Press.
- Lalonde MLM, Marcus JM. 2017. The complete mitochondrial genome of the long-horned caddisfly Triaenodes tardus (Insecta: Trichoptera: Leptoceridae). Mitochondrial DNA B Resour. 2:765–767.
- Liao F, Wang L, Wu S, Li Y-P, Zhao L, Huang G-M, Niu C-J, Liu Y-Q, Li M-G. 2010. The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci. 6:172–186.
- Living Prairie Mitogenomics Consortium. 2017. The complete mitochondrial genome of the lesser aspen webworm moth Meroptera pravella (Insecta: Lepidoptera: Pyralidae). Mitochondrial DNA B Resour. 2:344–346.
- Marcus JM. 2018. Our love-hate relationship with DNA barcodes, the Y2K problem, and the search for next generation barcodes. AIMS Genetics. 5:1–23.
- Marcus JM, Hughes TM, McElroy DM, Wyatt RE. 2010. Engaging first year undergraduates in hands-on research experiences: the Upper Green River barcode of life project. J Coll Sci Teach. 39:39–45.
- McCullagh BS, Marcus JM. 2015. The complete mitochondrional genome of Lemon Pansy, Junonia lemonias (Lepidoptera: Nymphalidae: Nymphalinae). J Asia-Pacific Ent. 18:749–755.
- Neave F. 1933. Ecology of two species of Trichoptera in Lake Winnipeg. Int Rev Ges Hydrobiol. 29:17–28.
- Peirson DSJ, Marcus JM. 2017. The complete mitochondrial genome of the North American caddisfly Anabolia bimaculata (Insecta: Trichoptera: Limnephilidae). Mitochondrial DNA B Resour. 2:595–597.
- Peters MJ, Marcus JM. 2017. Taxonomy as a hypothesis: testing the status of the Bermuda buckeye butterfly Junonia coenia bergi (Lepidoptera: Nymphalidae). Syst Ent. 42:288–300.
- Selgeby JH. 1974. Immature insects (Plecoptera, Trichoptera, and Ephemeroptera) collected from deep-water in western Lake Superior. J Fish Res Bd Can. 31:109–111.
- Sievers F, Wilm A, Dineen D, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Syst Biol. 7:539.
- Swofford DL. 2002. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, Massachusetts, USA: Sinauer Associates.
- Wang Y, Liu X, Yang D. 2014. The first mitochondrial genome for caddisfly (Insecta: Trichoptera) with phylogenetic implications. Int J Biol Sci. 10:53–63.
- Williams DD, Penak BL. 1980. Some aspects of case building in Phryganea cinerea Walker (Trichoptera, Phryganeidae). Anim Behav. 28:103–110.
- Winter WD. 2000. Basic techniques for observing and studying moths and butterflies. Vol 5. Los Angeles, CA: The Lepidopterists’ Society.
