Abstract
The whole chloroplast (cp) genome sequence of Musella lasiocarpa has been characterized from Illumina pair-end sequencing. The complete cp genome was 169,178 bp in length, containing a large single copy (LSC) region of 87,884 bp and a small single copy (SSC) region of 11,144 bp, which were separated by a pair of 35,075 bp inverted repeat (IR) regions. The genome contained 138 genes, including 88 protein-coding genes (87 PCG species), 37 tRNA genes (30 tRNA species), and eight ribosomal RNA genes (four rRNA species). The most of gene species occur as a single copy, while 23 gene species occur in double copies. The overall AT content of M. lasiocarpa cp genome is 63.3%, while the corresponding values of the LSC, SSC, and IR regions are 64.9, 69.2, and 60.3%, respectively. The cp genome sequence is similar to that of the genus Musa.
Musella lasiocarpa (Franch.) C. Y. Wu ex H. W. Li, a medicinal plant, is the only species in the monotypic genus Musella of the Musaceae. It is distributed in Guizhou, Sichuan, and Yunnan provinces in China. The fresh flowers and bracts can be used for medicine to stop bleeding and counteract inflammation. This medicinal plant is widely used to treat enteritis, constipation and female diseases, detoxify monkshood poisoning and alleviate drunkenness (Liu and Kress 2003). However, due to anthropogenic over-exploitation and decreasing distributions, this species needs urgent conservation. Knowledge of the genetic information of this species would contribute to the formulation of protection strategy. In this study, we assembled and characterized the complete chloroplast (cp) genome sequence of M. lasiocarpa based on the Illumina pair-end sequencing data ().
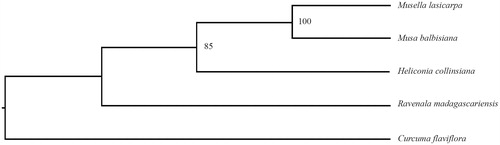
Figure 1. Phylogenetic relationships of Musaceae species using whole chloroplast genome. GenBank accession numbers: C. flaviflora (Nc_028729.1), M. balbisiana (Nc_028439.1), H. collinsiana (Nc_020362.1), and R. madagascariensis (Nc_022927.1).
Fresh leaves of M. lasiocarpa were collected from Dayao (Chuxiong, Yunnan, China; coordinates: 101°14′33″E, 25°54′10″N). Total genomic DNA was extracted with a modified CTAB method (Doyle and Doyle Citation1987). First, we obtained 10 million high quality pair-end reads for M. lasiocarpa and retained cp genome reads by mapping reads to all published Solanaceae cp genomes using BWA v0.7.12 (Li and Durbin Citation2009) and SAMtools v1.2 (Li et al. Citation2009). Second, we assembled these reads into a complete cp genome using Velvet v1.2.07 and Geneious v8.1.4 (Zerbino and Birney Citation2008; Kearse et al. Citation2012). Third, we annotated the plastid genomes using Plann v1.1 (Huang and Cronk Citation2015) and corrected the annotation with Geneious v8.1.4 (Kearse et al. Citation2012) and Sequin v13.70 (http://www.ncbi.nlm.nih.gov/Sequin/). A neighbour-joining (NJ) tree with 100 bootstrap replicates was inferred using TreeBeST 1.9.2 (Albert et al. Citation2009). The complete cp genome sequence was deposited in GenBank under accession number KY807173.
The M. lasiocarpa cp genome is 169,178 bp in length, exhibits a typical quadripartite structural organization, consisting of a large single copy (LSC) region of 87,884 bp, two inverted repeat (IR) regions of 35,075 bp and a small single copy (SSC) region of 11,144 bp. The cp genome contains 138 complete genes, including 88 protein-coding genes (87 PCGs), eight ribosomal RNA genes (four rRNAs), and 37 tRNA genes (30 tRNAs). Most genes occur in a single copy, while 23 genes occur in double, including all rRNAs (4.5S, 5S, 16S, and 23S rRNA), seven tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and 12 PCGs (rps7, rps12, rps15, rps19, rpl2, rpl23, ndhA, ndhB, ndhH, ycf1, ycf15, and ycf68). The overall AT content of cp DNA is 63.3%, while the corresponding values of the LSC, SSC, and IR regions are 64.9%, 69.2%, and 60.3%, respectively. Curcuma flaviflora was used as an outgroup, phylogenetic analysis of three plastid genomes from published species of Solanaceae indicated that the M. lasiocarpa clustered together with Musa balbisiana, and then formed one clade with Heliconia collinsiana in the Musaceae.
In summary, the complete cp genome from this study not only provides important insight into conservation and restoration efforts for M. lasiocarpa, but also plays a critical role in constructing phylogeny of the Musaceae family.
Acknowledgements
The authors are grateful to the opened raw genome data from public database.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Albert JV, Jessica S, Abel UV, Li H, Richard D, Ewan B. 2009. EnsemblCompara GeneTrees: Complete duplication-aware phylogenetic trees in vertebrates.. Genome Res. 19:327–335.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform.. Bioinformatics. 25:1754–1760.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis B, Durbin R. 2009. 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools.. Bioinformatics. 25:2078–2079.
- Liu AZ, Kress JK. 2003. The ethnobotany of Musella lasiocarpa (Musaceae), an endemic plant of southwest China. Econ Bot. 57:279–281.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
