Abstract
Graphomya rufitibia (Diptera: Muscidae) was distributed worldwide. In this study, the complete mitogenome of G. rufitibia was sequenced and annotated, and the full-length was a 15,374 bp fragment, consisting of A (40.64%), G (8.96%), T (36.80%), and C (13.59%). The mitogenome is composed of 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and a non-coding AT-rich region. Most PCGs used the canonical putative codon to start and stop, 21 tRNAs are folded into the classic clover-leaf structure, with the one exception of tRNA-Ser (AGN). A total length of 11,182 bp encoding 3727 amino acids, and account for 72.7% of the whole mitogenome. Phylogenetic analyses showed that G. rufitibia belongs to family Mydaeinae.
Graphomya rufitibia mainly spread over Asia and Australia, the larvae were semi-aquatic (Collin Citation1948; Arntfield Citation1975). It was reported that adult and larvae were coprophagous (Huang et al. Citation2007), thus bionomics of G. rufitibia making it was detrimental in animal husbandry and public health. Adults can be identified by the characteristic, but it is hard to distinguish in its young age, species identification by mitogenome is fast, economical, and reliable. More and more molecular studies about Muscidae have been published to supplement traditional taxonomy as rapid species identification ways.
Specimens of G. rufitibia were captured in August 2017 in Changsha city (28°12′N, 112°58′E), China. All voucher specimens were assigned a unique field code and reserved in the Guo’s Lab (Changsha city). The G. rufitibia mitogenome has been submitted to GenBank with accession number MG735216.
The complete mitogenome of G. rufitibia was amplified with 11 overlapping short PCR primers. PCR amplicons were conducted with the following procedures: initial denaturation at 94 °C for 4 min, followed by 35 cycles of 30 s at 94 °C, annealed at 50 °C for 30 s, and elongation 1 min at 72 °C, end in a final elongation for 10 min at 72 °C. PCR products were using an ABI PRISM 3730 automated sequencer (Applied Biosystems, Foster, CA) to sequence. The primitive sequence files were manually corrected for repeats at the beginning and end of the sequence to authorize a circular mitogenome, and then assembled into contigs with BioEdit 7.0.9.0 (Hall Citation1999). All encoding regions were manually aligned and validated. Open reading frames (ORFs) were executed through the NCBI ORF Finder according to the mitochondrial genetic codes of invertebrate. To make sure the authenticity of gene boundaries, all PCGs were compared with other muscid mitochondrial sequences under performed in MEGA 7.0 (Kumar et al. Citation2016).
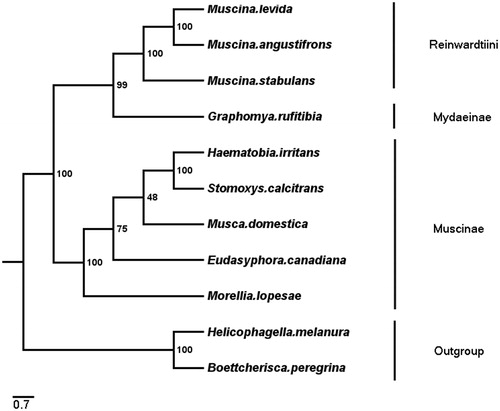
Phylogenetic analyses of G. rufitibia with eight Muscidae species were performed based on 13 PCGs +2 rRNAs by using ML methods (), and used two Sarcophagidae species as outgroups. Phylogenetic tree shows two clades, clade 1 consists of Reinwardtiini (Muscina genus) and Mydaeinae (Graphomya genus), which was corresponding to Kutty et al. (Citation2014) research (Kutty et al. Citation2014). The clade 2 consists of Muscinae (Stomoxyini+ Muscini), the topological structure was congruent with the previous studies (Kutty et al. Citation2014; Haseyama et al. Citation2015).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Arntfield PW. 1975. A revision of graphomya robineau-desvoidy (Diptera: Muscidae) from North America. Can Entomol. 107:257–302.
- Collin JE. 1948. On the classification of the genera allied to Musca L. (Diptera). Syst Entomol. 17:125–127.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Haseyama KLF, Wiegmann BM, Almeida EAB, de Carvalho CJB. 2015. Say goodbye to tribes in the new house fly classification: a new molecular phylogenetic analysis and an updated biogeographical narrative for the Muscidae (Diptera). Mol Phylogenet Evol. 89:1–12.
- Huang YT, Shinonaga S, Sasaki H. 2007. Studies on the muscid flies associated with pasturing cattle and water buffaloes in Taiwan (Diptera: Muscidae). J Rakuno Gakuen Univ. 32(1):15–20.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Kutty SN, Pont AC, Meier R, Pape T. 2014. Complete tribal sampling reveals basal split in Muscidae (Diptera), confirms saprophagy as ancestral feeding mode, and reveals an evolutionary correlation between instar numbers and carnivory. Mol Phylogenet Evol. 78:349–364.