Abstract
Blowfly species of the family Calliphoridae can be used in forensic investigations to estimate the minimum post-mortem interval (PMImin). Lucilia caesar and Lucilia illustris (Diptera: Calliphoridae) are closely related and phenotypically similar, making reliable identification difficult. To identify potential genetic markers to distinguish these species, five complete mitochondrial genomes were sequenced: three for L. caesar (KM657111–KM657113) and two for L. illustris (KM657109, KM657110). The ND6 gene contained the most species-specific SNPs (1.71%), followed by the ND5 gene (1.68%) and the COI gene (1.56%), identifying ND6 and ND5 as valuable loci for differentiating L. caesar and L. illustris specimens.
Species from the family Calliphoridae are typically the first to colonize a corpse (Rodriguez and Bass Citation1983) and their development can be used to determine the time since oviposition (Amendt et al. Citation2007), and hence an estimation of the PMImin, which can aid legal investigations (Easton and Smith Citation1970; Erzincllioglu Citation1983; Kulshrestha and Chandra Citation1987). The two closely related greenbottles Lucilia caesar and Lucilia illustris are both members of the family Calliphoridae and are commonly found in the UK. These species are very similar and morphological identification is a challenging task even for experienced entomologists (Stevens and Wall Citation1996; Wallman and Donnellan Citation2001).
mtDNA-based identification has been explored in previous studies, most frequently looking at COI, to distinguish species, including L. caesar and L. illustris (Malgorn and Coquoz Citation1999; Wells and Sperling Citation2000; Wallman Citation2001; Harvey et al. Citation2003; Sonet et al. Citation2012). The COII gene, the 16S ribosomal gene and the cyt b gene also showed insufficient interspecific variation between L. caesar and L. illustris (Sonet et al. Citation2012).
The samples were all collected from a site in Burnley UK (53.7661140 N, −2.2366190 W), which is at an altitude of 268 m. DNA was extracted using the DNeasy® Blood and Tissue Kit (Qiagen, Hilden, Germany). The genomes of five specimens were amplified with 21 custom-made primer pairs and Sanger sequenced (KM657109.1, KM657110.1, KM657111.1, KM657112.1, KM657113.1). DNA extracts are stored at the University of Central Lancashire, UK.
Genes were identified using ORF Finder (Wheeler et al. Citation2002), tRNAscan-SE (v 1.21) (Lowe and Eddy Citation1996), and alignment with other complete Lucilia mitochondrial genomes (L. sericata AJ422212, L. porphyrina JX913758, L. cuprina JX913744) (Stevens et al. Citation2008; Nelson et al. Citation2012). Newly sequenced mitochondrial genomes of both species consisted of 13 protein-coding genes, 22 tRNA genes and two rRNA genes: 23 are located on the heavy strand (tRNA-Ile, tRNA-Met, ND2, tRNA-Trp, COI, tRNA-Leu(UUA), COII, tRNA-Lys, tRNA-Asp, ATP8, ATP6, COIII, tRNA-Gly, ND3, tRNA-Ala, tRNA-Arg, tRNA-Asn, tRNA-Ser(AGC), tRNA-Glu, tRNA-Thr, ND6, cyt b, and tRNA-Ser(UCA)). The other 14 genes are located on the light strand. This genome organization is consistent with other Lucilia species AJ422212, JX913758, and JX913744 (Stevens et al. Citation2008; Nelson et al. Citation2012).
Start and stop codons were also similar to other Diptera Elodia flavipalpis (Zhao et al. Citation2013) and Chrysomya chloropyga (Junqueira et al. Citation2004): the most used start codon was ATG (6), but ATT (4) and ATA (2) were also present; the COI start codon is TCG (serine), which is not a typical start codon, but has been observed in other Diptera (including L. cuprina and L. sericata) – the ATTTAA sequence adjacent to the start codon is involved in translation initiation (Junqueira et al. Citation2004). Incomplete termination codons (T) were observed for COII, ND4, and ND5; it is thought that the termination codon is completed by polyadenylation (Lessinger et al. Citation2000).
Two extra copies of tRNA-Leu and tRNA-Ser were found, which is typical for most Diptera. Some Diptera contain more duplicate copies of tRNAs, but this was not the case for L. caesar and L. illustris (Lessinger et al. Citation2000; Harvey et al. Citation2003; Junqueira et al. Citation2004; Duarte et al. Citation2008). Anti-codons of the tRNAs were identified and showed to be consistent with other Diptera (L. sericata AJ422212, L. porphyrina, JX913758, L. cuprina JX913744). Based on tRNAscan-SE results and RNAfold (Lorenz et al. Citation2011), all identified tRNAs successfully folded into typical cloverleaf structures, except for tRNA-Ser(AGC), which missed the D stem. However, this has been observed in other species as well, including Diptera (Junqueira et al. Citation2004).
The length of the ribosomal RNAs, 1327 bp for the 16S subunit and 787 bp for the 12S subunit was the same as other Lucilia species (L. sericata AJ422212, L. porphyrina JX913758, and L. cuprina JX913744). The control regions of L. caesar and L. illustris were 1122 bp and 1121 bp, respectively; this is shorter than L. sericata (1125 bp) and L. cuprina (1136), but larger than L. porphyrina (1047 bp).
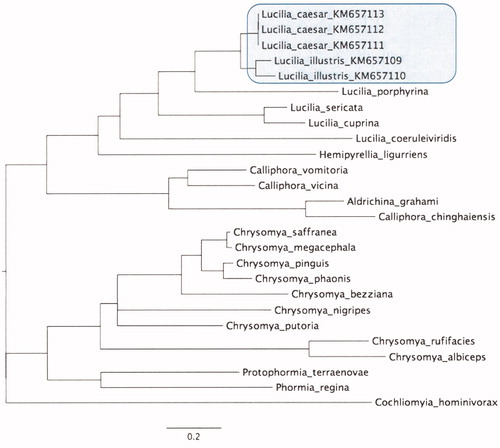
Besides finding possible new genetic markers, the complete mitochondrial genomes provide insight into the evolution of Calliphoridae. To infer phylogenetic relationships among Calliphoridae, we performed Bayesian phylogenetic analysis based on complete mitochondrial genome sequences (). PartitionFinder (Lanfear et al. Citation2012) was run prior to phylogenetic analysis to find the most supported data partitioning and associated substitution models based on the Bayesian Information Criterion (BIC). Fourteen partitions were identified for the alignment of the complete mitochondrial genomes of analysed Calliphoridae. All tested Lucilia species formed a monophyletic group. Phylogenetic analysis confirmed L. illustris and L. caesar as sister species. There were two distinct clades among tested Calliphoridae species, one included Lucilia spp., Calliphora spp., Hemipyrellia sp., Aldrichina sp. while the other clade included Chrysomya spp., Protophormia sp., and Phormia sp. A polytomy was present in the final tree, represented by Cochliomyia sp.
Figure 1. The Calliphoridae (NC_019636, NC_026668, NC_009733, NC_019637, KM657109-KM657113, NC_019573, NC_029486, NC_019638, NC_002660, NC_019635, NC_019634, NC_002697, NC_025338, NC_031381, NC_028412, NC_019633, NC_019632, NC_019631, NC_028411, NC_019639, NC_029215, NC_026996) phylogenetic summary tree inferred using the Bayesian approach implemented in MrBayes 3.2 (Ronquist et al. Citation2012) with Sarcophagidae: Sarcophaga crassipalpis (NC_026667) and Ravinia pernix (NC_026196) as an outgroup (not shown). Mitogenomes sequenced in this study are highlighted. All nodes are well-supported (PP > 0.9). Four independent runs were performed for 10 million generations and sampled every 1000 generations. Convergence of replicates was verified in TRACER v. 1.6 (Rambaut et al. Citation2014) by ensuring unimodality of posterior distributions and effective sample size (ESS >200) for all parameters. Trees were summarized using the 50% majority rule method, after discarding the first 25% of the sample as burnin, and visualized using FIGTREE 1.4.2 (Rambaut Citation2012).
Most studies for genetic identification of L. caesar and L. illustris have been based on the COI gene. Other mitochondrial genes that have been used are the COII gene, the 16S ribosomal gene and the cyt b gene, but these show insufficient interspecific variation between these two Calliphoridae indicating that additional genetic markers need to be developed (Sonet et al. Citation2012). In this study, the ND6 and ND5 genes contained the most species specific SNPs (1.71% and 1.68%, respectively), followed by the COI gene (1.56%). Although these results are based on only few mitochondrial genomes, the results are consistent with the study of Nelson et al. (Citation2012), where the ND6 gene showed the highest interspecific genetic variation within Calliphoridae (and Chrysomyinae), when compared to other mitochondrial genes. Further work, including population studies, is needed to evaluate the value of ND6 and NR5 as markers for differentiating between L. caesar and L. illustris in forensic casework.
Disclosure statement
The authors report no conflict of interest and are responsible for the content and writing of this article. The University of Central Lancashire’s Ethics Committee approved all experimental procedures.
References
- Amendt J, Campobasso CP, Gaudry E, Reiter C, Leblanc HN, Hall M. 2007 . Best practice in forensic entomology-standards and guidelines. Int J Legal Med. 121:90–104.
- Duarte GT, De Azeredo-Espin AML, Junqueira ACM. 2008. The mitochondrial control region of blowflies (Diptera: Calliphoridae): a hot spot for mitochondrial genome rearrangements. J Med Entomol. 45:667–676.
- Easton AM, Smith KGV. 1970. The entomology of the cadaver. Med Sci Law. 10:208–215.
- Erzincllioglu YZ. 1983. The application of entomology to forensic medicine. Med Sci Law. 23:57–63.
- Harvey ML, Dadour IR, Gaudieri S. 2003. Mitochondrial DNA cytochrome oxidase I gene: potential for distinction between immature stages of some forensically important fly species (Diptera) in Western Australia. Forensic Sci Int. 131:134–139.
- Junqueira ACM, Lessinger AC, Torres TT, da Silva FR, Vettore AL, Arruda P, Azeredo-Espin AML. 2004. The mitochondrial genome of the blowfly Chrysomya chloropyga (Diptera: Calliphoridae). Gene. 339:7–15.
- Kulshrestha P, Chandra H. 1987. Time since death. An entomological study on corpses. Am J Forensic Med Pathol. 8:233–238.
- Lanfear R, Calcott B, Ho SYW, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Lessinger AC, Martins Junqueira AC, Lemos TA, Kemper EL, da Silva FR, Vettore AL, Arruda P, Azeredo-Espin AML. 2000. The mitochondrial genome of the primary screwworm fly Cochliomyia hominivorax (Diptera: Calliphoridae). Insect Mol Biol. 9:521–529.
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. 2011. ViennaRNA Package 2.0. Algorithms Mol Biol. 6:26.
- Lowe TM, Eddy SR. 1996. TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Malgorn Y, Coquoz R. 1999. DNA typing for identification of some species of Calliphoridae. An interest in forensic entomology. Forensic Sci Int. 102:111–119.
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 511:131–142.
- Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v1.6; [accessed 2017 Dec 20]. http://beast.bio.ed.ac.uk/Tracer/
- Rambaut A. 2012. Figtree v1.4.2; [accessed 2017 Dec 20]. http://tree.bio.ed.ac.uk/software/figtree/
- Rodriguez WC, Bass WM. 1983. Insect activity and its relationship to decay rates of human cadavers in East Tennessee. J Forensic Sci. 28:423–432.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. Mrbayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Sonet G, Jordaens K, Braet Y, Desmyter S. 2012. Why is the molecular identification of the forensically important blowfly species Lucilia caesar and Lucilia illustris (family Calliphoridae) so problematic? Forensic Sci Int. 223:153–159.
- Stevens J, Wall R. 1996. Classification of the genus Lucilia (Diptera: Calliphoridae): a preliminary parsimony analysis. J Nat Hist. 30: 1087–1094.
- Stevens JR, West H, Wall R. 2008. Mitochondrial genomes of the sheep blowfly, Lucilia sericata, and the secondary blowfly, Chrysomya megacephala. Med Vet Entomol. 22:89–91.
- Wallman JF, Donnellan SC. 2001. The utility of mitochondrial DNA sequences for the identification of forensically important blowflies (Diptera: Calliphoridae) in southeastern Australia. Forensic Sci Int. 120:60–67.
- Wallman JF. 2001. Third-instar larvae of common carrion-breeding blowflies of the genus Calliphora (Diptera: Calliphoridae) in South Australia. Invertebr Syst. 15:37–51.
- Wells JD, Sperling FA. 2000. A DNA-based approach to the identification of insect species used for postmortem interval estimation and partial sequencing of the cytochrome oxydase b subunit gene I: a tool for the identification of European species of blow flies for postmortem interval estimation. J Forensic Sci. 45:1358–1359.
- Wheeler DL, Church DM, Lash AE, Leipe DD, Madden TL, Pontius JU, Schuler GD, Schriml LM, Tatusova TA, Wagner L, et al. 2002. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 30:13–16.
- Zhao Z, Su T, Chesters D, Wang S, Ho SYW, Zhu C, Chen X, Zhang C. 2013. The mitochondrial genome of Elodia flavipalpis Aldrich (Diptera: Tachinidae) and the evolutionary timescale of Tachinid flies. PLoS One. 8:e61814.
