Abstract
The complete chloroplast genome of Sisymbrium irio was determined. The length of the complete chloroplast genome is 154,001 bp. The whole chloroplast genome consists of 83,891 bp long single copy (LSC) and 17,630 bp small single copy (SSC) regions, separated by a pair of 26,240 bp inverted repeat (IR) regions. The S. irio chloroplast genome encodes 112 annotated known unique genes including 79 protein-coding genes, 30 tRNA genes, and four rRNA genes. The phylogenetic position of S. irio is sister to Brassiceae and Thlaspideae.
Keywords:
Sisymbrium irio belongs to the tribe Sisymbrieae in the family Brassicaceae. The tribe Sisymbrieae is phylogenetically close to the tribe Brassiceae, included in the Lineage II (Bailey et al. Citation2006; German et al. Citation2009; Beilstein et al. Citation2010; Couvreur et al. Citation2010; Warwick et al. Citation2010; Huang et al. Citation2016). Sisymbrium irio, known as London Rocket, is a common weed and used for analyses of repetitive sequences and comparative studies of tribe Brassiceae (Grellet et al. Citation1989; Hall et al. Citation2011). Due to its production of a large number of seeds, this species had been naturalized worldwide as invasive species and are found often abundant. Draft genome sequence of S. irio was already determined for comparative genomic analysis (Haudry et al. Citation2013).
A strain sampled from Pakistan (ABRC accession CS22563, SASSC J04) was used for sequencing. Chloroplast was isolated as described in Okegawa and Motohashi (Citation2015). DNAs were extracted from the isolated chloroplasts by DNeasy Plant Mini kit (Qiagen, Hilden, Germany) and sequenced as the single-ended reads using the NextSeq500 platform (Illumina Co., San Diego, CA). The generated reads were assembled by velvet 1.2.10 (Zerbino and Birney Citation2008) and were also mapped to Sinapis arvensis whole chloroplast genome sequence (GenBank accession # KU050690) using bowtie (Langmead and Salzberg Citation2012) to construct complete chloroplast genome sequence. The average depth of chloroplast genome was 845.7. The nucleotide sequence was submitted to DDBJ (accession number: LC375846).
The complete chloroplast genome of S. irio has a total length of 154,001 bp, consisting of 83,891 bp LSC region and 17,630 bp SSC region separated by a pair of 26,240 bp inverted repeat (IR) regions. This structure is identical to those of other species in Brassicaceae. The overall GC content is 36.32% and the GC contents of the LSC, SSC, and IR regions are 34.09%, 29.24%, and 42.26%, respectively. There are 86 protein coding genes, 37 tRNA genes, and eight rRNA genes typical for the chloroplast genome of Brassicaceae. Most genes occur as a single copy, except for seven protein coding, seven tRNA, and four rRNA genes within the IR regions are duplicated in whole chloroplast genome. Sometimes, a few chloroplast genes were lost in many plant species (Guo et al. Citation2017), however, S. irio has complete set of protein coding genes in Brassicaceae species without any pseudogenization.
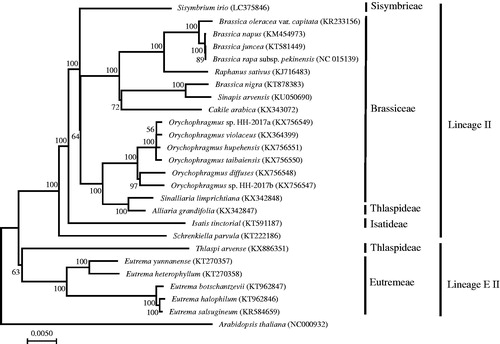
The phylogenetic tree including S. irio and species in Lineage II and EII was constructed by complete chloroplast genomes (). 76 protein coding gene sequences were concatenated to estimate NJ tree with JC corrected synonymous divergence by MEGA ver. 7 (Kumar et al. Citation2016). Sisymbrium irio was sister to a clade containing Brassiceae and Thlaspideae species as reported by phylogenetic studies with partial chloroplast and nuclear sequences (Beilstein et al. Citation2010; Couvreur et al. Citation2010), whereas nuclear ITS regions showed different clustering where Sisymbrieae clustered with Isatideae and Schizopetaleae (German et al. Citation2009; Warwick et al. Citation2010). The complete chloroplast genome sequence of S. irio can provide a reference for other Sisymbrium species including many invasive weeds.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bailey CD, Koch MA, Mayer M, Mummenhoff K, O’Kane SL Jr, Warwick SI, Windham MD, Al-Shehbaz IA. 2006. Toward a global phylogeny of the Brassicaceae. Mol Biol Evol. 23:2142–2160.
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. 2010. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci USA. 107:18724–18728.
- Couvreur TL, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. 2010. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol Biol Evol. 27:55–71.
- German DA, Friesen N, Neuffer B, Al-Shehbaz I, Hurka H. 2009. Contribution to ITS phylogeny of the Brassicaceae, with special reference to some Asian taxa. Plant Syst Evol. 283:33–56.
- Grellet F, Delcasso-Tremousaygue D, Delseny M. 1989. Isolation and characterization of an unusual repeated sequence from the ribosomal intergenic spacer of the crucifer Sisymbrium irio. Plant Mol Biol. 12:695–706.
- Guo X, Liu J, Hao G, Zhang L, Mao K, Wang X, Zhang D, Ma T, Hu Q, Al-Shehbaz IA, Koch MA. 2017. Plastome phylogeny and early diversification of Brassicaceae. BMC Genomics. 18:176.
- Hall JC, Tisdale TE, Donohue K, Wheeler A, Al-Yahya MA, Kramer EM. 2011. Convergent evolution of a complex fruit structure in the tribe Brassiceae (Brassicaceae). Am J Bot. 98:1989–2003.
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. 2013. An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet. 45:891–898.
- Huang CH, Sun R, Hu Y, Zeng L, Zhang N, Cai L, Zhang Q, Koch MA, Al-Shehbaz I, Edger PP, et al. 2016. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol Biol Evol. 33:394–412.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
- Okegawa Y, Motohashi K. 2015. Chloroplastic thioredoxin m functions as a major regulator of Calvin cycle enzymes during photosynthesis in vivo. Plant J. 84:900–913.
- Warwick SI, Mummenhoff K, Sauder CA, Koch MA, Al-Shehbaz IA. 2010. Closing the gaps: phylogenetic relationships in the Brassicaceae based on DNA sequence data of nuclear ribosomal ITS region. Plant Syst Evol. 285:209–232.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.