Abstract
Cuscuta pentagona is a parasitic plant whose seeds are often mixed with the seeds of medicinal Cuscuta species. To facilitate the identification of C. pentagona seeds, we generated the complete chloroplast genome sequence of C. pentagona Engelm. using the Illumina MiSeq platform. The complete chloroplast genome is 86,380 bp long, with a 50,958 bp LSC region, a 7022 bp SSC region, and two inverted repeat (IRa and IRb) regions comprising 14,200 bp. The chloroplast genome consists of 85 unique genes, 57 protein-coding genes, four ribosomal RNA (rRNA) genes, and 24 transfer RNA (tRNA) genes. Two gene families, NADH oxidoreductases and RNA polymerase-related genes, are absent in this genome. Phylogenetic analysis revealed that C. pentagona is closely related to C. reflexa and C. exaltata, with strong support values.
Cuscuta pentagona Engelm., a member of the Convolvulaceae family, is widely distributed worldwide (Hwang et al. Citation2013). Cuscuta pentagona originated in North America, is a parasitic plant that grows by attaching itself to its host plant and winding around the host via thin, leafless yellow stem with a diameter of approximately 1 mm. This well-known plant parasitizes various hosts and causes extensive crop damage (Alakonya et al. Citation2012). Although the dried seeds of other Cuscuta species are utilized as valuable medicinal plant materials, frequently, these seeds are indiscriminately mixed with C. pentagona seeds in the Korean herbal market (KIOM Citation2016). Thus, we sequenced the chloroplast genome of C. pentagona to enable species identification and to protect the herbal medicine market.
Fresh vines were collected from C. pentagona plants grown in medicinal plantations in Korea (36°24′56.2″ N 127°23′26.1″ E). The C. pentagona plants were given identification numbers, and specimens were registered in the Korean Herbarium of Standard Herbal Resources (Index herbariorum code KIOM) at the Korea Institute of Oriental Medicine (KIOM) under Voucher no. KIOM201701018523. Genomic DNA was extracted from the samples using a DNeasy Plant Maxi kit (Qiagen, Valencia, CA). An Illumina paired-end sequencing library was constructed and generated using the MiSeq platform (Illumina Inc., San Diego, CA). High-quality paired-end reads of approximately 2.1 Gb were obtained from C. pentagona. The chloroplast contigs of C. pentagona were de novo assembled from low-coverage whole-genome sequences.
The complete chloroplast genome of C. pentagona is 86,380 bp in length, relatively small compared to other angiosperms (GenBank Accession no. MH121054). The C. pentagona chloroplast genome has a typical quadripartite structure consisting of a large single copy (LSC) region of 50,958 bp, a small single copy (SSC) region of 7022 bp, and a pair of inverted repeats (IRa and IRb) comprising 14,200 bp. The GC content of the C. pentagona chloroplast genome is 37.9%, with the IR regions having higher GC content (43.2%) than the LSC (36%) and SSC (29.6%) regions. The C. pentagona chloroplast genome contains 85 unique genes, including 57protein-coding genes, 24 tRNAs, and 4 rRNAs. Among these, 12 genes were duplicated in the IR regions. The chloroplast genome of C. pentagona harbours genes related to photosynthesis but lacks NADH and RNA polymerase genes (Revill et al. Citation2005). A few tRNA genes are absent in this genome.
To investigate the phylogenetic relationship of C. pentagona with other plant species, we aligned 56 protein-coding gene sequences shared by the 20 other taxa. We generated a ML tree containing 13 nodes with 100% bootstrap support values (). The phylogenetic relationship is well supported in the Convolvulaceae and Solanaceae families. The ML tree indicates that C. pentagona forms a sister relationship with C. reflexa and C. exaltata in the Convolvulaceae family, with 100% bootstrap support values ().
Figure 1. Maximum Likelihood tree based on the chloroplast protein-coding genes of 21 taxa including C. pentagona and Oryza sativa as outgroup. Fifty-nine protein-coding genes were aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate the Maximum Likelihood phylogenetic tree with MEGA 6.0 (Tamura et al., Citation2013). The bootstrap support values (>50%) from 1000 replicates are indicated in the nodes.
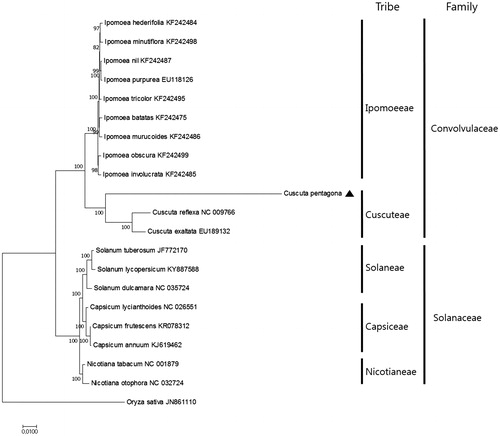
These findings should facilitate the development of tools that can be used to identify C. pentagona to help protect the herbal medicine market.
Acknowledgments
We thank the Classification and Identification Committee of the KIOM for identifying plant materials and the Herbarium of Korea Standard Herbal Resources (Index herbariorum code KIOM) for providing plant materials.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Alakonya A, Kumar R, Koenig D, Kimura S, Townsley B, Runo S, Garces HM, Kang J, Yanez A, David-Schwartz R, et al. 2012. Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. Plant Cell. 24:3153–3166.
- Hwang S, Kil J, Lee C-W, Kim Y. 2013. Distribution and host plants of parasitic weed Cuscuta pentagona Engelm. Hangug Jaweon Sigmul Haghoeji. 26:289–302.
- [KIOM] Korea Institute of Oriental Medicine. 2016. Defining Dictionary for Medicinal Herbs. [accesed 2018 Mar 3] http://boncho.kiom.re.kr/codex/
- Revill MJ, Stanley S, Hibberd JM. 2005. Plastid genome structure and loss of photosynthetic ability in the parasitic genus Cuscuta. J Exp Bot. 56:2477–2486.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729.
