Abstract
Chrysanthemum is an important ornamental, herbal, and medicinal plant. We report the complete mitochondrial genome (mitogenome) sequence of Chrysanthemum boreale. The mitogenome is 211,002 bp in length, has a GC content of 45.36%, and contains 58 genes, including 35 protein-coding genes, three ribosomal RNA genes, and 20 transfer RNA genes. A phylogenetic analysis based on mitogenome protein sequences from various plants confirmed that C. boreale belongs to the Asteraceae family. This mitogenome will be useful in evolutionary and phylogenetic studies of Chrysanthemum and Asteraceae.
Chrysanthemum, a genus of flowering plants in the Asteraceae family, is famous for its diversity in flower shape, size, and colour and includes ornamental, commercial, and wild species. Chrysanthemum boreale, a wild species with small, yellow flowers, is distributed throughout eastern Asia (Kim et al. Citation2003) and has been used for medicinal and herbal purposes, due to its antibacterial, anti-inflammatory, and skin-regenerating properties (Kim et al. Citation2003; Kim et al. Citation2010; Kim et al. Citation2015). As C. boreale is resistant to white rust disease, this plant may be a suitable genetic resource to develop disease-resistant chrysanthemum cultivars (Park et al. Citation2014). Only two mitogenomes from the Asteraceae family (Helianthus annuus and Diplostephium hartwegii) have been deposited in GenBank and no mitogenome for the Chrysanthemum genus has been reported. In this study, we report the construction of the complete C. boreale mitogenome. This mitogenome will be useful in evolutionary and phylogenetic studies of Chrysanthemum and Asteraceae.
To construct the complex and dynamic mitogenome of C. boreale (GenBank BioSample SAMN07296937), long-read sequences were generated and assembled using PacBio’s Single Molecule Real-Time (SMRT) platform. The plant was collected from the Korea (N 35° 29′ 00″ E 126° 48′ 00″) and kept in National Institute of Horticultural and Herbal Science, Rural Development Administration with the ID of IT121002 (Hwang et al. Citation2013). A library was prepared from total genomic DNA using a SMRTbell Template Prep Kit 1.0 (Pacific Biosciences, PN 100-259-100) and sequenced on PacBio’s RS II platform using P6-C4 chemistry (DNAlink, Republic of Korea). The obtained raw reads were primarily assembled with the FALCON and FALCON-Unzip algorithms (Chin et al. Citation2016). Mitogenome-like sequences were searched by BLASTn against the complete mitogenome of H. annuus (GenBank Accessino number: CM007908.1), assembled into a single contig by CANU (Version 1.4), with the genome size set at 400,000, and finally circularized using MUMmer (Kurtz et al. Citation2004; Koren et al. Citation2017). The mitogenome was annotated using MITOFY and manually compared with other mitochondrial protein sequences (Alverson et al. Citation2010).
The C. boreale mitogenome was deposited in GenBank under accession number MH004292. The mitogenome is 211,002 bp in length, has a G+C content of 45.36%, and is predicted to contain 58 genes, including 35 protein-coding genes, three ribosomal RNA genes, and 20 transfer RNA genes. While most protein-coding genes had an ATG as the start codon, three (atp6, ccmFn, and nad4L) had an ACG, rendering them susceptible to C-to-U mRNA editing, which is common in plant mitochondria (Giegé and Brennicke Citation1999). Additionally, mttB had ATT as the start codon, which was commonly observed in H. annuus (Grassa et al. Citation2016). Three NADH dehydrogenase subunit genes, nad1, nad2, and nad5, are trans-spliced and seven genes (nad4, nad7, cox2, ccmFc, rps1, rps3, and rps14) contain introns, whereas the remaining 48 genes consist of a single exon.
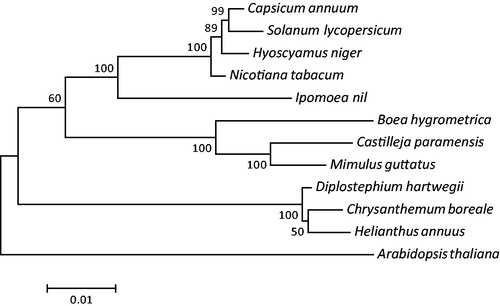
Phylogenetic analysis was conducted using the mitogenomes of 12 plant species with Arabidopsis thaliana as an outgroup. The maximum likelihood (ML) tree based on the translated amino acid sequences of 20 common protein-coding genes (atp1, atp4, atp8, atp9, ccmB, ccmC, ccmFn, cob, cox1, cox3, matR, mattB, nad3, nad4, nad4L, nad6, nad9, rpl5, rps4, and rps12) showed that C. boreale and other Asteraceae species (H. annuus and D. hartwegii) form a monophyletic group supported by the bootstrap value of 100% (). This complete mitogenome sequence of C. boreale is the first reported in the Chrysanthemum genus and represents a useful genetic resource to identify C. boreale and to conduct phylogenetic and evolutionary studies.
Figure 1. Maximum-likelihood phylogenetic tree based on the concatenated amino acid sequences of 20 protein-coding genes in 12 plant mitogenomes. Arabidopsis thaliana was used as an outgroup. Numbers at the nodes indicate bootstrap values for 1000 replicates. All the mitogenome sequences are available in GenBank: Chrysanthemum boreale (MH004292), A. thaliana (Y08501), Boea hygrometrica (NC_016741), Capsicum annuum (KJ865410), Castilleja paramensis (NC_031806), Diplostephium hartwegii (NC_034354), Helianthus annuus (NC_023337), Hyoscyamus niger (NC_026515), Ipomoea nil (NC_031158), Mimulus guttatus (NC_018041), Nicotiana tabacum (NC_006581), and Solanum lycopersicum (NC_035963).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27:1436–1448.
- Chin C-S, Peluso P, Sedlazeck FJ, Nattestad M, Concepcion GT, Clum A, Dunn C, O'Malley R, Figueroa-Balderas R, Morales-Cruz A, et al. 2016. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 13:1050.
- Giegé P, Brennicke A. 1999. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc Natl Acad Sci. 96:15324–15329.
- Grassa CJ, Ebert DP, Kane NC, Rieseberg LH. 2016. Complete mitochondrial genome sequence of sunflower (Helianthus annuus L). Genome Announc. 4:e00981–e00916.
- Hwang Y-J, Younis A, Bok RK, Lim K-B, Eun C-H, Lee J, Sohn S-H, Kwon S-J. 2013. Karyomorphological analysis of wild Chrysanthemum boreale collected from four natural habitats in Korea. Flower Res J. 21:182–189.
- Kim K-J, Kim Y-H, Yu H-H, Jeong S-I, Cha J-D, Kil B-S, You Y-O. 2003. Antibacterial activity and chemical composition of essential oil of Chrysanthemum boreale. Planta Med. 69:274–277.
- Kim Y, Sung J, Sung M, Choi Y, Jeong H-S, Lee J. 2010. Involvement of heme oxygenase-1 in the anti-inflammatory activity of Chrysanthemum boreale Makino extracts on the expression of inducible nitric oxide synthase in RAW264.7 macrophages. J Ethnopharmacol. 131:550–554.
- Kim DY, Won K-J, Yoon M-S, Hwang DI, Yoon SW, Park J-H, Kim B, Lee HM. 2015. Chrysanthemum boreale Makino essential oil induces keratinocyte proliferation and skin regeneration. Nat Prod Res. 29:562–564.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27:722–736.
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12.
- Park SK, Lim JH, Shin HK, Jung JA, Kwon YS, Kim MS, Kim KS. 2014. Identification of Chrysanthemum genetic resources resistant to white rust caused by Puccinia horiana. Plant Breed Biotechnol. 2:184–193.
