Abstract
The mitogenome of the Siberian flying squirrel Pteromys volans is a circular molecule of 16,514 bp, consisting of a control region and a conserved set of 37 genes containing 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes (12S rRNA and 16S rRNA). The mitogenome of the Korean P. volans is AT-biased, with a nucleotide composition of 32.2% A, 30.4% T, 12.5% G, and 24.8% C. The phylogenetic analysis revealed that P. volans is well placed within the tribe Pteromyini (Sciuridae: Sciurinae), which forms a sister clade to the flying squirrels of the genus Petaurista.
The Siberian flying squirrels of Pteromys volans (Sciuridae), also known as the Old World flying squirrel, are distributed across a wide range from the Baltic Sea in the west to the Pacific Coast in the east, including the Korean Peninsula and northeast China (Shar et al. Citation2016). In South Korea, P. volans was designated as endangered species as well as natural monument.
We sequenced and characterized the complete mitogenome of a P. volans individual caught in Gangneung, South Korea. The voucher specimen (KNPS00027853) was deposited in the National Park Research Institute of Korea National Park Service. Genomic DNA extraction, PCR and gene annotation were conducted according to the previous studies (Jeon and Park Citation2015; Rahman et al. Citation2016). A previously published mitogenome of the P. volans (JQ230001) was used as reference for gene annotation using Geneious 8.0.5 (Kearse et al. Citation2012). Phylogenetic tree was constructed under the General Time Reversible model (GTR + R+I) (Nei and Kumar Citation2000) using maximum-likelihood (ML) procedures implemented in MEGA6 (Tamura et al. Citation2013). Glis glis (NC001892) was used as outgroup.
The complete mitogenome (MH212330) of the P. volans contains 16,514 bp in length, which consists of a control region (CR) and a conserved set of 37 vertebrate mitochondrial genes including 13 PCGs, 22 tRNA genes, and 2 rRNA genes (12S rRNA and 16S rRNA). The order and orientation of these genes are identical to those of other mammals (Kim and Park Citation2016; Kim et al. Citation2017; Lim et al. Citation2017).
The mitogenome of the Korean P. volans is AT-biased, with a nucleotide composition of 32.2% A, 30.4% T, 12.5% G, and 24.8% C. Total length of the 13 mitochondrial PCGs of the Korean P. volans is 11,367 bp long (62.1% A+T), with the exclusion of stop codons, which encode 3789 amino acids. Lengths of two rRNA genes (12s rRNA and 16s rRNA) were 966 bp (61.4% A+T) and 1565 bp long (64.6% A+T), respectively. There are a total of 22 tRNA genes for transferring 20 amino acids, including include two leucine-tRNA genes (tRNALeu(UUR) and tRNALeu(CUN)) and two serine-tRNA genes (tRNASer(AGY) and tRNASer(UCN)). The mitochondrial OR is 30 bp long and is located between tRNAAsn and tRNACys in the WANCY region. Mitochondrial CR, which is located between tRNAPro and tRNAPhe, is 1067 bp long (62.5% A+T).
In intraspecific comparison with the previous published mitogenome of the P. volans (JQ230001) which has total size of 16,513 bp, insertion of only a base pair was found in mitochondrial CR. Mitogenomes of the two P. volans were identical in 99.1% of whole sites of the aligned sequences (16,517 sites). Identical sites of 95.9% were found in mitochondrial CR and pairwise identity of 99.3% was found in the other mitochondrial regions except for mitochondrial CR.
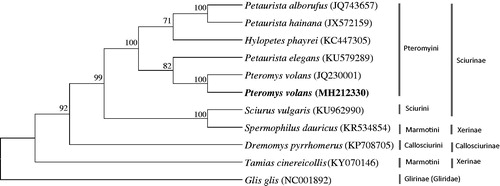
The phylogenetic analysis revealed that P. volans is well placed within the tribe Pteromyini (Sciuridae: Sciurinae), which forms a sister clade to the genus Petaurista (). The two tribes of Sciurini (Sciurus vulgaris) and Marmotini (Spermophilus dauricus) formed a sister clade, which is a sister to the tribe Pteromyini.
Figure 1. The phylogenetic relationship of Pteromys volans and its allied species inferred from maximum-likelihood analysis based on complete mitogenome sequences. The ML tree was generated using the GTR + G+I model, and the robustness of the tree was tested with 1000 bootstrap. The numbers on the branches indicate bootstrap values.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Jeon MG, Park YC. 2015. The complete mitogenome of the wood-feeding cockroach Cryptocercus kyebangensis (Blattodea: Cryptocercidae) and phylogenetic relations among cockroach families. Anim Cells Syst. 19:432–438.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kim HR, Jeon MG, Min JH, Kim HJ, Park YC. 2017. Complete mitochondrial genome of the roe deer Capreolus pygargus tianschanicus (Cervidae) from Korea. Mitochondrial DNA B Resour. 2:558–559.
- Kim HR, Park YC. 2016. Intraspecific comparison of two complete mitogenome sequences from the Korean water deer (Cervidae: Hydropotes inermis argyropus). Mitochondrial DNA A. 27:4101–4103.
- Lim SJ, Kim HR, Cho JY, Park YC. 2017. Complete mitochondrial genome of the least weasel Mustela nivalis (Mustelidae) in Korea. Mitochondrial DNA B Resour. 2:740–741.
- Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. New York: Oxford University Press.
- Rahman MM, Yoon KB, Kim JY, Hussin MZ, Park YC. 2016. Complete mitochondrial genome sequence of the Indian pipistrelle Pipistrellus coromandra (Vespertilioninae). Anim Cells Syst. 20:86–94.
- Shar S, Lkhagvasuren D, Henttonen H, Maran T, Hanski I. 2016. Pteromys volans (errata version published in 2017). The IUCN Red List of Threatened Species 2016: e.T18702A115144995; [accessed 2018 Apr 10]. http://dx.doi.org/10.2305/IUCN.UK.2016-3.RLTS.T18702A22270935.en
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
