Abstract
The mitogenome of Sellanucheza jaegeri was 15,623 bp long, revealed the same gene order to that of typical Polydesmida. Both the tRNASer(AGN) and tRNASer(UCN) lacked the DHU arms. No tandem repeat was found in two control regions. Phylogenetic analysis indicated that Sphaerotheriida was so antiquity that divided out earlier than others. We supported that Polydesmida had a relatively systematic affinity between Julida and Playtdesmida, and suggested that the interordinal phylogenetic relationships within Diplopoda should be further investigated.
Diplopoda (millipedes) is the third most diverse class of Arthropoda, have diversified for over 400 million years, with ∼12,000 species described and an estimated diversity of 80,000 living species (Zhang Citation2011; Brewer et al. Citation2012; Enghoff et al. Citation2015). They occupy an important niche in the ecosystem because of their function in the breakdown of organic matter. The rearrangement of genes in mitogenomes had remarkably occurred in Diplopoda. However, relationships among the Myriapoda remain unresolved in Robertson et al. (Citation2015), and owning to the lack of species sequences, the interordinal phylogenetic relationships within Diplopoda are one of the most debated issues.
Sample (voucher no. MP06) of Sellanucheza jaegeri was collected from Feng County (34°12'45''N, 106°54'09''E), Shaanxi, China. Genomic DNA was prepared in paired-end libraries, tagged and subjected to the Illumina Xten platform (Illumina, San Diego, CA) with 150 bp paired-end strategy, and yield 16,147,682 paired-end raw reads. Mapping against the complete mitogenome of Appalachioria falcifera (GenBank: JX437063), high-quality reads were assembled using MITObim version 1.9 (University of Oslo, Norway) (Hahn et al. Citation2013). A total of 201,061 individual mitochondrial reads gave an average coverage of 1879.4X. Comparing with the Asiomorpha coarctata (GenBank: KU721885), annotations were generated in MITOchondrial genome annotation Server (MITOS) (Bernt et al. Citation2013) and Geneious version 11.0.4 (Biomatters Ltd, New Zealand).
The complete mitogenome sequence consist of 15,623 bp for S. jaegeri (Genbank: MH213061) and with the base composition A + T is 61.4%. Its genes’ arrangement and orientation are matched known Polydesmida mtDNA pattern (Dong et al. Citation2016). The typical ATG start codon is present in all PCGs except for ND1 and ND2 being with ATA and TTG, respectively. Two types of stop codons TAA and TAG are used for most genes, with the exception of incomplete stop codon T for ATP6, ND1, and ND5.
The two rRNA genes, 692 bp in srRNA and 1248 bp in lrRNA, are located between control region 1 (CR1) and tRNALeu(CUN) and separated by tRNAVal. All the tRNA genes have typical cloverleaf secondary structures except for the tRNASer(AGN) and tRNASer(UCN), which lack the DHU arms. The 1017 bp long CR1 is located between tRNASer(UCN) and srRNA, and the 181 bp long CR2 are located between tRNAThr and tRNAHis, respectively. Of the genome sequenced here, no tandem repeat was found in these two regions.
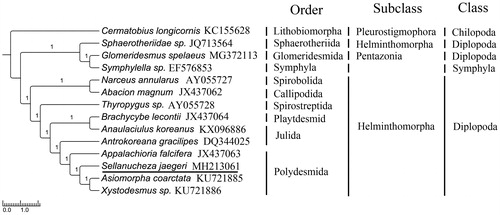
To validate the phylogenetic analyses of S. jaegeri (Polydesmida), MrBayes (Ronquist et al. Citation2012) and RAxML (Stamatakis Citation2006) were used to reconstruct BI and ML tree, undering the best partitioned scheme and optimal model analyzed in Partitionfinder (Lanfear et al. Citation2012) (Model GTR + I + G). Cermatobius longicornis (Genbank: KC155628) and Symphylella sp. (Genbank: EF576853) were selected as out-groups. The phylograms obtained from BI and ML (data not shown) were all strongly indicated that Sphaerotheriida with an ancestral gene order of mitogenome for the Helminthomorpha was located in the basal branch (Gai et al. Citation2008; Dong et al. Citation2012). The topological structures between Spirobolida, Callipodida, Spirostreptida, Playtdesmida, and Julida were similar with that observed for other previously studies (Woo et al. Citation2007; Brewer et al. Citation2013). Extraordinarily, Julidae and Nemasomatidae were not clustered together, what is more, Xystodesmidae and Paradoxosomatidae were not a sister group in this study . These results provide novel molecular information that can potentially be used for phylogenetic of Diplopoda ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Brewer MS, Sierwald P, Bond JE. 2012. Millipede taxonomy after 250 years: classification and taxonomic practices in a mega-diverse yet understudied arthropod group. PLoS One. 7:e37240.
- Brewer MS, Swafford L, Spruill CL, Bond JE. 2013. Arthropod phylogenetics in light of three novel millipede (myriapoda: diplopoda) mitochondrial genomes with comments on the appropriateness of mitochondrial genome sequence data for inferring deep level relationships. PLoS One. 8:e68005.
- Dong Y, Xu JJ, Hao SJ, Sun HY. 2012. The complete mitochondrial genome of the giant pill millipede, Sphaerotheriidae sp. (Myriapoda: Diplopoda: Sphaerotheriida). Mitochondrial DNA. 23:333–335.
- Dong Y, Zhu LX, Bai Y, Ou YY, Wang CB. 2016. Complete mitochondrial genomes of two flat-backed millipedes by next-generation sequencing (Diplopoda, Polydesmida). Zookeys. 637:1–20.
- Enghoff H, Golovatch SI, Short M, Stoev P, Wesener T. 2015. Diplopoda-taxonomic overview. In: Minelli A, editor. Treatise on zoology-anatomy, taxonomy, biology. the myriapoda, Vol. 2, Chapter 2. Leiden, the Netherlands: Brill; p. 363–453.
- Gai YH, Song DX, Sun HY, Yang Q, Zhou KY. 2008. The complete mitochondrial genome of Symphylella sp. (Myriapoda: Symphyla): extensive gene order rearrangement and evidence in favor of Progoneata. Mol Phylogenet Evol. 49:574–585.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. Partitionfinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701.
- Robertson HE, Lapraz F, Rhodes AC, Telford MJ. 2015. The complete mitochondrial genome of the Geophilomorph Centipede Strigamia maritima. PLoS One. 10:e0121369.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinform. 22:2688–2690.
- Woo HJ, Lee YS, Park SJ, Lim JT, Jang KH, Choi EH, Choi YG, Hwang UW. 2007. Complete mitochondrial genome of a troglobite millipede Antrokoreana gracilipes (Diplopoda, Juliformia, Julida), and juliformian phylogeny. Mol Cells. 23:182–191.
- Zhang ZQ. 2011. Phylum arthropoda von siebold, 1848. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Zootaxa. 4138: 99–103.