Abstract
Taxonomic ambiguity exists in genus Systomus and recently many new species were described under this genus. Systomus sarana subnasutus is considered a valid subspecies of S. sarana sarana although revisions have been done by some researchers. We employed a combination of morpho-meristics and molecular tools (Cytochrome c oxidase I, 16S and Cytochrome b genes of mitochondrial genome) to resolve the two species. Three morpho-meristic characters, head length/maxillary barbel length (HL/MxBL), Lateral Line Scales (LLSs) as well as two truss-based characters, had discernible variation between the two taxa. The sequence analysis (2353 nucleotides) depicted a separate clad of S. sarana subnasutus with high bootstrap support. The findings from combined use of morphology, meristics and mitogenes were concordant. The corroborative results suggest the possibility of two different species. The results suggest to adopt suitable management measures, accordingly.
Introduction
Genus Systomus (subfamily: Cyprininae; family: Cyprinidae; order: Cypriniformes) is an economically important group, comprising of 19 fish species native to tropical Asia (Kottelat Citation2013). Systomus sarana sarana (Hamilton Citation1822) commonly known as olive barb is widely distributed in south-east Asian countries. It has wider occurrence throughout India except peninsular region and south to Krishna River (Talwar and Jhingran Citation1991). Recent study (Dahanukar Citation2010) indicated reduction in its natural abundance due to anthropogenic pressures, and places it under vulnerable group. The another subspecies, S. sarana subnasutus, popularly known as peninsular olive barb, is endemic to the Western Ghats (Dahanukar et al. Citation2004) and inhabits the river Krishna and all other rivers south to it (Menon 1963, Citation1999). Availability of this species in different rivers of peninsular India has also been reported by several other authors (Chandanshive et al. Citation2007; Jadhav and Yadav Citation2009; Shahnawaz and Venkateshwarlu Citation2009).
There have been several revisions for this species. Historically, Valenciennes first described S. sarana subnasutus as Barbus subnasutus from Pondicherry, India (Cuvier and Valenciennes Citation1842). Menon (Citation1963) synonymized the species with Puntius sarana but considered it as a valid subspecies. Later on, this was considered Barbodes sarana subnasutus by Menon (Citation1999). While cataloguing, Eschmeyer and Fricke (Citation2011) again synonymized this species with P. sarana, and Jayaram (2010) considered it as a valid subspecies. Dahanukar (Citation2013) had strongly recommended for further taxonomic investigation, especially using molecular markers.
The morphology of fishes has been the key source for taxonomic studies. However, the current trend advocates the potential role of molecular tools to support morphological inferences in resolving ambiguities, especially among the closely related species. Mitochondrial genes are the proven tools to infer variability at inter-/intra-species level, depending upon their evolutionary rates. The slow evolving genes such as COI, 16S have been widely used for inter-species relatedness. In the present study, a combined use of morpho-meristics and three mitogenes was done to resolve taxonomic ambiguity between S. sarana sarana and its peninsular congener subspecies, S. sarana subnasutus.
Materials and methods
Fish specimens, used in present study, were collected from commercial catches of various rivers between December 2014 to September 2015 (). The collected specimens were identified using standard taxonomic keys (Talwar and Jhingran Citation1991). Digital images were captured for truss morphometry (Karaoglu et al. Citation2011), at site. For each specimen, a total of 23 traditional morphometric measurements and 10 meristic characters were recorded. A total of 14 landmarks, covering the entire shape of individual specimens, yielded 91 inter-landmarks. The truss distances were extracted using tpsDig2 v2.1 (Rohlf Citation2006) and PAST (Hammer et al. Citation2001).
Table 1. Specimen information, collection localities and accession number of Systomus sarana sarana and Systomus sarana subnasutus (N is the number of samples analysed).
Genetic analysis
Gene sequencing
Total gDNA was extracted from individual muscle tissue samples using the phenol–chloroform method. The protocol of Ruzzante et al. (Citation1996) with slight modification (Singh et al. Citation2012) was followed. The amplification reaction was set in ABI Veriti thermocycler. The universal primers were used for amplifying Cyt b (Xiao et al. 2001); 16S rRNA (Palumbi et al. 1991) and COI (Ward et al. 2005). The amplicons were sequenced bi-directionally on ABI 3730 sequencer.
Data analysis
Statistical analysis
To determine inter-specific variations, traditional morphometric and meristic characters were used separately in analysis as their allocation abilities are different, statistically (Karaoglu and Belduz Citation2011). Species were compared using ratios among various traditional morphometric characters and mean values of meristic characters. A one-way analysis of variance (ANOVA) was conducted on each of the variables for principal component analysis (PCA) and discriminant function analyses (DFA).
The raw DNA sequences were edited and aligned using BioEdit software version 7.0.5.2 (Hall Citation1999) and Clustal W (Thompson et al. Citation1997). A total of 655 bp (COI), 557 (16S) and 1141 (Cyt b) were analysed. The sequence characteristics, such as polymorphic sites, nucleotide composition, and transition/transversion ratios were determined.
To ascertain the inter-relatedness of the two subspecies of S. sarana, gene sequences of S. orphoides, S. chalakkudiensis and S. denisonii were downloaded from NCBI GenBank and analysed together. Neighbour joining (NJ) tree was generated with 1000 replications (Saitou and Nei Citation1987).
Results
Morpho-meristics and Truss network
The ANOVA revealed eight morphometric ratios and four meristic characters to exhibit significant (p < .05) differences between the two taxa. However, discriminant analysis of significant variables demonstrated three variables namely, head length/maxillary barbel length (HL/MxBL), lateral line scales (LLSs) and vertebrae counts (VCs). Significant (p < .05) Fisher’s distances between the groups were observed.
The ANOVA revealed that the samples of two species differed significantly (p < .05) at 57 transformed morphometric characters, out of a total 90. PCA extracted 10 principal components accounting for 92.2% of the total variation. HL/MxBL, LLSs, VCs, eye diameter and distance between pectoral fin origin to operculum differentiated the two Systomus species.
Molecular analysis
The multiple alignment of COI gene (655 bp) from two species, S. sarana sarana (19) and S. sarana subnasutus (18), revealed six haplotypes, with two and four haplotypes were found, respectively (). A total of 636 sites (97.1%) were conserved 19 (2.9%) variable and 17 (2.6%) parsimony informative. The analysis depicted the average nucleotide frequencies as A = 27.9%, T = 28.4%, G = 17.2% and C= 26.5% which showed higher AT (56.3%) than GC content (43.7%). The order of occurrence of bases was T > A > C > G in both the species. Overall, the occurrence of transitional events was more commonly observed than transversions.
Table 2. Haplotypes and variable sites of COI sequences.
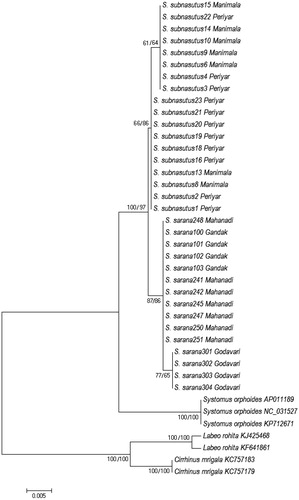
The genetic distance within populations (for both taxa) did not exhibit significant differences. However, the average genetic distance between the S. sarana subnasutus and S. sarana sarana were 2.4% and 2.5%, respectively (). NJ tree was constructed (K2P distance) using all 37 COI sequences including congeners from NCBI. Monophyly of Systomus (Sahayadria) group was presented when Indian major carp species were used as outgroups. However, the polyphyletic clustering was obvious for Systomus and Sahayadria genera. The topology is similar to our own work (under review) which supported the distinctiveness of three taxa (). Furthermore, strong bootstrap support indicated the divergence of Systomus genera with that of Sahayadria which is in line with conventional taxonomy. The tree shows 100% bootstrap to support distinction between two taxa, S. sarana sarana and S. sarana subnasutus.
Figure 1. Phylogenetic relationships of Systomus species with close relatives based on K2P divergence of COI gene.
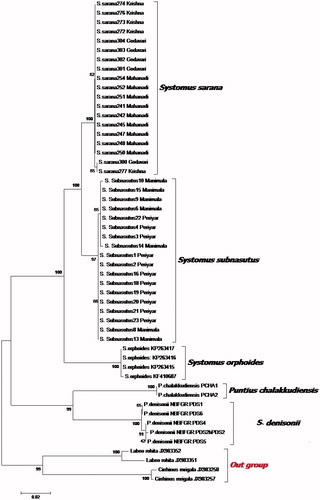
Table 3. Genetic distance between S. sarana subnasutus and S. sarana sarana.
Sequence analysis of 16S rRNA (557 bp) revealed two haplotypes, each from S. sarana sarana (n = 15) and S. sarana subnasutus (n = 18). The genetic distance was 0.18% within taxa, while 0.5% observed between individuals of S. sarana sarana and S. sarana subnasutus. Analysis of Cytochrome b (1141 bp) presented 1090 conserved sites, whereas 51 were variable ones. Pairwise genetic distance between S. sarana sarana and S. sarana subnasutus was 3.73%, whereas different populations of both the species showed low genetic distance (0.89% and 0.35%), respectively. The NJ tree highlighted similar tree topologies with high bootstrap (71–100) () for both mtDNA markers.
Discussion
Previously, several researchers demonstrated the taxonomic status of both species (Menon Citation1999; Jayaram Citation2010; Eschmeyer and Fricke Citation2011). The genus Puntius has been considered one of the largest group among subfamily cyprininae. This subfamily is represented by over 120 valid species, widely distributed in south and south-east Asia. With all these revisions, the species of genus Puntius (Hamilton Citation1822) along with P. sophore as type species are recognized under the six distinct genera Puntius, Systomus, Dawkinisia, Haludaria (Dravidia), Sahyadria and Pethia. Sarana group of genus Systomus is a complex consisting of S. sarana sarana and its three valid subspecies viz. S. sarana spilurus, S. sarana subnasutus and S. sarana orphoides.
Traditional morphology is often used for species identification by gathering and analysing, data from large sample sizes, though molecular markers are proven to be more robust in reconstructing phylogenies. The combined use of the morphological characters and mitochondrial data provides a strong framework for discriminating species, compared to the use of morphological characters alone. In the present investigation, we used molecular evidences (along with morphological attributes) to corroborate the taxonomic status of two species.
The overall results from this study indicated that S. sarana sarana differs significantly from S. sarana subnasutus. Earlier, Jayaram (Citation1991), in his study on genus Puntius reported that in P. sarana sarana (Hamilton), LLSs are 30–34 with mostly 31 or 32; and all populations are distributed north of the Krishna river system in Southern India, while its subspecies, P. sarana subnasutus (Val.), the LLSs range 28–31 and all populations were distributed in Krishna river (and south to it) in the Peninsular India. In P. sarana sarana contains a dark blotch on lateral line before base of caudal fin which is distinguishable with the similar blotch at 24th scale in P. sarana subnasutus (Jayaram Citation2010).
In S. sarana sarana, eye diameter is larger than S. sarana subnasutus. Similarly, distance between pectoral fin origin to operculum is larger in S. sarana sarana compared to S. sarana subnasutus. In this study, truss morphometry also yielded concordant results. Truss-based differentiation has been used in several fish species (Cavalcanti et al. Citation1999; Parsons et al. Citation2003; Sakai et al. Citation2009). The analysis of 2353 nucleotides, further confirmed the possibility of differentiation between S. sarana sarana and S. sarana subnasutus on the basis of NJ phylogram which was supported by high bootstrap values in all three mitochondrial markers. The pattern of clustering demonstrated their separate existence.
Conclusively, with the molecular evidence corroborating to the morphological inferences, this study strongly advocates that there is probability that S. sarana subnasutus is a valid species and distinct from the S. sarana sarana. However, taxonomic re-description is required for elevating from subspecies to species level. The findings are important for conservation and management of resources.
Acknowledgements
The present work was accomplished under a project entitled, ‘Phylogeography and Genetic Stock Structure of Olive barb, Systomus sarana sarana (Hamilton Citation1822) Population using Mitochondrial and Nuclear DNA Markers’. The authors are thankful to Department of Science and Technology, Government of India for funding support. Simultaneously, excellent technical assistance provided by Sh. R.S. Sah is duly acknowledged.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Cavalcanti MJ, Monteiro LR, Lopes PRD. 1999. Landmark-based morphometric analysis in selected species of serranid fishes (percifomes: Teleostei). Zool Stud. 38:287–294.
- Chandanshive NE, et al. 2007. Fish fauna of Pavanariver Pune. Zoos' Print J. 22:2693–2694.
- Cuvier G, Valenciennes A. 1842. Histoire naturelle des poissons. Tome seizième. P. Bertrand, Paris, 472 p.
- Dahanukar N, Raut R, Bhat A. 2004. Distribution, endemism and threat status of freshwater fishes in the Western Ghats of India. J Biogeography. 31:123–136. IUCN. 2011. IUCN Red List of Threatened Species (ver. 2011.1).
- Dahanukar N. 2010. Labeo dyocheilus. The IUCN Red List of Threatened Species 2010: e.T166625A6249964. http://dx.doi.org/10.2305/IUCN.UK.2010-4.RLTS. T166625A 6249964.en. Downloaded on 05 July 2016.
- Dahanukar N. 2013. Systomus sarana ssp. subnasutus. The IUCN Red List of Threatened Species 2013: e.T172514A6907469. http://dx.doi.org/10.2305/IUCN.UK.2011-1.RLTS.T172514A6907469.en downloaded on 17.05.2013.
- Eschmeyer WN, Fricke R, editors. 2011. Catalog of fishes. Updated internet version of 30 September 2011. Catalog databases of CAS cited in FishBase (website).
- Hall TA. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98.
- Hamilton F. 1822. An account of the fishes found in river Ganges and its branches. Edinburgh and London. p. i–vii 1–405, Pls.1–39.
- Hammer Ø, Harper DAT, Ryan PD. 2001. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica Data Analysis. Palaeontologia Electronica. 4:4–9.
- Jadhav SS, Yadav BE. 2009. A note on the ichthyofauna of Solapur District, with first report of a cyprinid fish Rasboracaverii (Jerdon) from Maharashtra State, India. J Threat Taxa. 1:243–244.
- Jayaram KC. 1991. Revision of the genus Puntius Hamilton from Indian region (Pisces: Cypriniformes, Cyprinidae, Cyprininae). Records of Zoological Survey of India. Occ. Paper: 135:52–58, 61–67, 113–122.
- Jayaram KC. 2010. The freshwater fishes of the Indian region, 2nd ed. Delhi: Narendra Publishing House; p. 616; 39 plates.
- Karaoglu H, Belduz AO. 2011. Multivariate discrimination among three Trachurus species from Turkey. J Anim Vet Adv. 10:121–127.
- Kottelat M. 2013. The fishes of the inland waters of Southeast Asia: a catalogue and core bibliography of the fishes known to occur in freshwaters, mangroves and estuaries. Raffles Bull Zool. (27):1–663.
- Menon AGK. 1963. Subspecies of the cyprinid fish Puntius sarana with description of P. saranaspilurus from Ceylon. Spolia Zeylanica. 30:65–70.
- Menon AGK. 1999. Check list - fresh water fishes of India. Records of the Zoological Survey of India. Misc Publ Occas. p. 366. Pap. No. 175.
- Palumbi SR, Benzie J. 1991. Large mitochondrial DNA differences between morphologically similar Penaeid shrimp. Mol Mar Biotechnol. 1:27–34.
- Parsons L, Harris KL, Turner K, Whitington PM. 2003. Roundabout gene family functions during sensory axon guidance in the Drosophila embryo are mediated by both Slit-dependent and Slit-independent mechanisms. Dev Biol. 264:363–375.
- Rohlf FJ. 2006. tpsDig2, Version 2. 1. State University of New York, Stony Brook. Available at: http://life.bio.sunysb.edu/morph
- Ruzzante DE, Taggart C, Cook D, Goddard S. 1996. Genetic differentiation between inshore and offshore Atlantic cod (Gadus morhua) off Newfoundland: microsatellite DNA variation and antifreeze level. Can J Fish Aquat Sci. 53:634–645.
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 4:406–425.
- Sakai H, Iguchi K, Yamazaki Y, Sideleva VG, Goto A. 2009. Morphological and mtDNA sequence studies on three crucian carps (Carassius: Cyprinidae) including a new stock from the Ob River system, Kazakhstan. J Fish Biol. 74:1756–1773.
- Shahnawaz A, Venkateshwarlu M. 2009. A checklist of fishes from the Tunga and Bhadra rivers, Karnataka, India with a special note on their biodiversity status. Current Biotica. 3:232–243.
- Singh RK, Lal KK, Mohindra V, Punia P, Sah RS, Kumar R, Gupta A, Das R, Lakra WS, Ayyappan S. 2012. Genetic diversity of Indian Major Carp, Labeo calbasu (Hamilton, 1822) populations inferred from microsatellite loci. Biochem. Syst Ecol. 44:307–316.
- Talwar PK, Jhingran AG. 1991. Inland fishes of India and adjacent countries. Volume 2. Rotterdam: A.A. Balkema.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgens DG. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 25:4876–4882.
- Valenciennes MA. 1842. Histoire Naturelle Poissons. Paris. 17:23–497.
- Ward RD, Tayler SZ, Bronwyn HI, Peter RL & Hebert PDN. 2005. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. doirstb.2005.1716/rstb.2005.1716
- Xiao WH, Zhang YP, Liu HZ. 2001. Molecular systematics of Xenocyprinae (Teleostei: Cyprinidae): Taxonomy, biogeography, and coevolution of a special group restricted in East Asia. Mol Phylogenet Evol. 18:163–173.