Abstract
The complete mitochondrial genome of Urocissa erythroryncha is 16930 bp in length. It was predicted to contain 13 PCGs, 22 tRNA genes, and 2 rRNA genes, and a putative control region. All of the PCGs initiated with ATG, except for MT-COX1 which began with GTG and MT-ND3 began with ATA, while stopped by three types of stop codons. Phylogenetic analysis showed that Urocissa erythroryncha and the other species of Corvidae were monophyletic group in this study. And the monophyly of the genus Pyrrhocorax was strongly supported. Moreover, our results also support a sister-group relationship between Corvidae and Muscicapidae.
Passeriformes is the largest order of Aves (Clements et al. 2016). The birds generally known as corvids is a widespread family named Corvidae (Aves: Passeriformes) (Gill and Donsker 2018). In our study, an adult Urocissa erythroryncha specimen was caught from Wuhu (geographic coordinate: 31.33°N, 118.38°E), Anhui Province, southeastern of China. The specimen was preserved in ethanol and stored to Anhui Normal University, under the voucher number Kan-K0024. Through the standard phenol/chloroform methods, the total genomic DNA of the Urocissa erythroryncha (code Kan-K0024) was extracted from the muscle tissue. The fourteen PCR and sequencing primers specific to U. erythroryncha mitochondrial genome were designed, and two long overlapping fragments were amplified to avoid the possibility of nuclear copies of mitochondrial genes (Kan et al. Citation2010; Zhang L et al. Citation2012; Zhang Q et al. Citation2017).
We obtained the complete mitochondrial genome of U. erythroryncha (GenBank Accession NC_020426) in this study. We described 16930 bp of U. erythroryncha mitochondrial genome DNA, including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and 1 putative control region. All tRNA genes can fold into a typical cloverleaf second structure. All of the PCGs initiated with ATG, except for MT-COX1 which began with GTG. And meanwhile three types of termination codons were identified. AGG for MT-COX1; TAA for MT-ND2, MT-COX2, MT-ATP6, MT-ATP8, MT-ND3, MT-ND4L, MT-ND6, and MT-CYTB; AGA for MT-ND1 and MT-ND5; and incomplete stop codon T- for MT-ND2, MT-COX3, and MT-ND4. We can detect one single control region (D-loop) between tRNAGlu and tRNAPhe. The D-loop region is 1346 bp in length. The overall base composition of the mitochondrial genome is as follows: A (30.92%), T (24.66%), G (14.32%), C (30.10%), with the A + T content of 55.58%. Nucleotide composition was calculated by MEGA v6.06 (Tamura et al. Citation2013). AT and GC skews were calculated by using the formulas (A – T)/(A + T) and (G – C)/(G + C) (Perna and Kocher Citation1995; Kan et al. 2016), respectively. The MT-ND6 gene of U. erythroryncha mitogenome has strong skews of T versus A (–0.41), and a strong skew of C versus G (0.52). The other 12 protein-coding genes of the U. erythroryncha mitochondrial genome have a slight skew of A vs T (0 to 0.20), and a strong skew of C versus G (GC skew = –0.75 to –0.30).
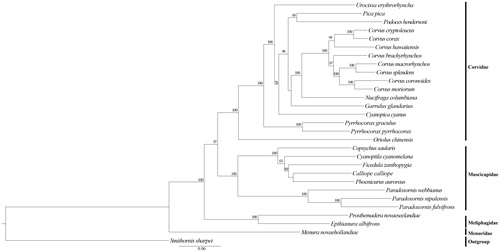
The phylogenetic position of U. erythroryncha was estimated from a concatenated dataset based on 13 PCGs. Maximum likelihood (ML) analyses were performed with Raxml GUI v 1.3.1 (Silvestro and Michalak Citation2012; Zhang et al. Citation2012; Jiang et al. Citation2015). The complete mitochondrial DNA sequences have been used successfully to estimate phylogenetic relationships among Passeriformes. The results showed that U. erythroryncha and the other species of Corvidae were monophyletic group in this study. Furthermore, other genera of Corvidae (Garrulus, Corvus, Pyrrhocorax, Nucifraga, Podoces, Pica, Cyanopica) were basal to the balance of the Corvidae. Moreover, our results also support a sister-group relationship between Corvidae and Muscicapidae. They all appear 100% bootstrap value in ML analyses which is strongly supported. We speculate that there is still a complex phylogenetic relationship between finch and other families that is not known to us. This study will contribute to the phylogenetic analyses in Passeriformes.
Figure 1. Phylogenetic tree of the relationships among Passeriformes based on the nucleotide dataset of the 13 PCGs. Smithornis sharpei was chosen as outgroup. ML analyses were implemented in ML + rapid bootstrap for 1000 replicates under GTRGAMMA, Branch lengths and topologies came from the ML analysis. All 29 species’ accession numbers are listed as below: Corvus brachyrhynchos NC_026461, C. corax NC_034838, C. cryptoleucus NC_034839, C. coronoides NC_035877, C. hawaiiensis NC_026783, C. macrorhynchos NC_027173, C. moriorum NC_031518, C. splendens NC_024607, Cyanopica cyanus NC_015824, Garrulus glandarius NC_015810, Nucifraga columbiana NC_022839, Oriolus chinensis NC_020424, Pica pica NC_015200, Podoces hendersoni NC_014879, Pyrrhocorax graculus NC_025927, P. pyrrhocorax NC_025926, Urocissa erythroryncha NC_020426, Epthianura albifrons NC_019664, Prosthemadera novaeseelandiae NC_029144, Menura novaehollandiae NC_007883, Calliope calliope NC_015074, Copsychus saularis NC_030603, C. cyanomelana NC_015232, Ficedula zanthopygia NC_015802, Paradoxornis nipalensis NC_028437, P. webbianus NC_024539, Phoenicurus auroreus NC_026066, Smithornis sharpei NC_000879.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Clements JF, Schulenberg ST, Iliff MJ, Sullivan BL, Wood CL, Roberson D. 2016. The eBird/Clements checklist of birds of the world:v2016. http://www.birds.cornell.edu/clementschecklist/download/.
- Gill F, Donsker D. 2018. IOC World Bird List (v 8.1). doi: 10.14344/IOC.ML.8.1.
- Jiang L, Chen J, Wang P, Ren Q, Yuan J, Qian C, Hua X, Guo Z, Zhang L, Yang J, et al. 2015. The mitochondrial genomes of Aquila fasciata and Buteo lagopus (Aves, Accipitriformes): sequence, structure and phylogenetic analyses. PLoS One. 10:e0136297.
- Kan X, Li X, Lei Z, Wang M, Chen L, Gao H, Yang Z. 2010. Complete mitochondrial genome of Cabot’s tragopan, Tragopan caboti (Galliformes: Phasianidae). Genet Mol Res. 9:1294–1216.
- Kan X, Ren Q, Ping W, Lan J, Zhang L, Ying W, Qin Z. 2014. Complete mitochondrial genome of the Eurasian siskin, Spinus spinus (Passeriformes: Fringillidae). Mitochondrial DNA A. 27:1–1835.
- Perna NT, Kocher TD. 1995. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J Mol Evol. 41:353–358.
- Silvestro D, Michalak I. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12:335–337.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Zhang Q, Wang Y, Chen R, Liu B, Kan X. 2017. The complete mitochondrial genome of Anas crecca (Anseriformes: Anatidae). Mitochondrial DNA B. 2:352–353.
- Zhang L, Wang L, Gowda V, Wang M, Li X, Kan X. 2012. The mitochondrial genome of the Cinnamon Bittern, Ixobrychus cinnamomeus (Pelecaniformes: Ardeidae): sequence, structure and phylogenetic analysis. Mol Biol Rep. 39:8315–8326.
