Abstract
Carpinus hebestroma (Betulaceae) is a critically endangered species endemic to Taiwan. In this study, we assembled the complete chloroplast (cp) genome of C. hebestroma and the cp genome was 159,231 bp in length, including a large single copy (LSC) region of 88,168 bp, a small single copy (SSC) region of 18,874 bp and a pair of inverted repeats (IRs) of 52,189 bp. The genome contained 124 genes, including 85 protein-coding genes, 30 tRNA genes and 8 rRNA genes. The overall GC content of the chloroplast genome was 36.46%. A phylogenetic analysis demonstrated a close relationship between C. hebestroma and Ostrya rehderiana.
Carpinus hebestroma Yamam is a species of small tree within the family Betulaceae, and this species was endemic to Taiwan and known only from the type specimen locality, in Batakan, Taiwan, China. Within this locality, the species has small and fragmented subpopulations, and the population was still declining because of the landslides after typhoons. This species was listed as critically endangered species in the IUCN red list now (http://www.iucnredlist.org/details/194577/0). Therefore, measures of conservation and restoration are urgently needed. The plastid genome will contribute to develop protection measures for this endangered species.
Fresh leaves were collected from an individual of C. hebestroma in Taiwan (24°09′17″ N, 121°29′33″ E; China; voucher 2015-Taiwan-001 deposited in the herbarium of Lanzhou University, Lanzhou, China). Genomic DNA was isolated using the modified CTAB method (Doyle Citation1987). The whole-genomic DNA data were sequenced using the Illumina Hiseq 2500 platform (Illumina, San Diego, CA). The paired-end data set comprised 72.2 million reads (2 × 150 bp). Chloroplast genome assembly using the Fast-Plast pipeline (https://github.com/mrmckain/Fast-Plast; McKain and Wilson, unpublished). Annotation was performed with Plann (Plastome Annotator, Huang and Cronk Citation2015).
The complete chloroplast genome of C. hebestroma (GenBank: MG720819) was 159,231 bp in length, comprising a LSC of 88,168 bp and a SSC of 18,874 bp, separated by IRs regions of 52,189 bp. It contained 124 genes, including 85 protein-coding genes, eight rRNA genes, and 30 tRNA genes. The plastome contained 95 unique genes, 14 genes duplicated in the IR regions. Among annotated genes, nine genes contained a single intron, and four genes had two introns (e.g. clpP, ycf3, rps12 and rpl2). The base compositions of major chloroplast genome were uneven (31.34% A, 18.60% C, 17.86% G and 32.20% T), with an overall GC content of 36.46%, and the corresponding values of the LSC, SSC and IR regions reaching 34.28%, 29.99% and 42.48%, respectively.
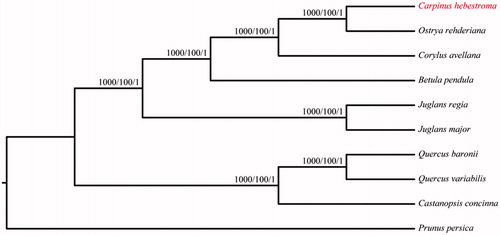
We also constructed the phylogenetic trees with Neighbour-Joining (NJ, Phylip version 3.696, Felsenstein Citation2005), the maximum likelihood (ML, RaxML version 8, Stamatakis Citation2014) and Bayesian analysis (BI, MrBayes version 3, Ronquist and Huelsenbeck Citation2003) methods based on the alignments created by the MAFFT (Katoh and Standley Citation2013) using 9 related species of Fagales and the Prunus persica as outgroup (). The results indicated that all three species from the Betulaceae were clustered together and C. hebestroma was most closely related to O. rehderiana. This complete chloroplast genome can be readily used for population genomic studies of C. hebestroma, and such information would be fundamental to formulate potential new conservation and management strategies for this endangered species.
Figure 1. The phylogenetic tree based on the 10 complete chloroplast genome sequences. NJ/ML/Bayesian posterior probabilities/bootstrap values are shown at nodes. Accession numbers: Betula pendula LT855378, Carpinus hebestroma MG720819, Castanopsis concinna NC_033409, Corylus avellana KX822768, Juglans major NC_035966, Juglans regia NC_028617, Ostrya rehderiana KT454094, Prunus persica HQ336405, Quercus baronii KT963087 and Quercus variabilis KU240009.
Disclosure statement
The authors report no potential conflicts of interest. The authors alone are responsible for content and writing of the paper.
Additional information
Funding
References
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences. Seattle: University of Washington, DC.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignmentsoftware version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
