Abstract
Stethojulis strigiventer is a tropical reef-associated marine fish belonging to the family Labridae. Herein, we report the first sequencing and assembly of the complete mitochondrial genome of S. strigiventer. The complete mitochondrial genome is 16,524 bp length and has the typical vertebrate mitochondrial gene arrangement, consisting of 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. Phylogenetic analysis using mitochondrial genomes of 11 species showed that Stethojulis strigiventer is clustered together with the genus Parajulis and Halichoeres and rooted with other Labridae species. This mitochondrial genome provides potentially important resources for addressing taxonomic issues of the Labriformes and studying conservation genetics.
The family Labridae, the wrasses, inhabit the tropical and subtropical waters of the Atlantic, Indian, and Pacific Oceans, though some species live in temperate waters (Bernardi et al. Citation2004; Rocha et al. Citation2010). The three-ribbon wrasse, Stethojulis strigiventer (Labriformes: Labridae), is listed as Least Concern in IUCN Red List due to the collection for the aquarium trade and habitat degradation in southeast Asia (Cabanban Citation2010). Although previous studies for many sub-groups of the Labridae obtained high nodal support (Hanel et al. Citation2002; Clements et al. Citation2004; Barber and Bellwood Citation2005), interrelationships among other familes such as Carapidae, Scaridae, Caesionidae, and Microdesmidae in Percomorpha are largely unresolved (Betancur-R et al. Citation2017). This is the first study to determine the complete mitochondrial genome of S. strigiventer, and to analyze the phylogenetic relationship of this species with members of Labridae.
The S. strigiventer specimen was collected from Chuuk ST 1, Micronesia (7.27N, 151.54E). Total genomic DNA was extracted from tissue of the specimen, which has been deposited in the National Marine Biodiversity Institute of Korea (Voucher No. MABIK 0000617). The mitogenome was sequenced and assembled using Illumina Hiseq 4000 sequencing platform (Illumina, San Diego, CA) and SOAPdenovo assembler at Macrogen Inc. (Korea), respectively. The complete mitochondrial genome was annotated using MacClade ver. 4.08 (Maddison and Maddison Citation2005) and DNASIS ver 3.2 (Hitachi Software Engineering).
The complete mitochondrial genome of S. strigiventer (GenBank accession no. AP018523) is 16,524 bp length, and includes 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a control region. The ND6 gene and eight tRNA genes are encoded on the light strand. The overall base composition of the heavy strand is 27.10% A, 28.83% C, 17.36% G, and 26.71% T. Similar to the mitogenomes of other vertebrates, the AT content is higher than the GC content (Saccone et al. Citation1999). All tRNA genes can fold into a typical cloverleaf structure, with lengths ranging from 66 to 74 bp. The 12S rRNA (952 bp) and 16S rRNA genes (1,693 bp) are located between tRNAPhe and tRNAVal and between tRNAVal and tRNALeu(UUR), respectively. Of the 13 protein-coding genes, 11 begin with an ATG start codon; the exception being the COI and ATP6 gene, which start with GTG and TTG, respectively. The stop codon of the protein-coding genes is TAA in COI, ATP8 and ND4L; TAG in NDI and ND6; AGA in ND5; TA in ATP6 and COIII; and T in the remaining five genes. The control region (836 bp) is located between tRNAPro and tRNAPhe.
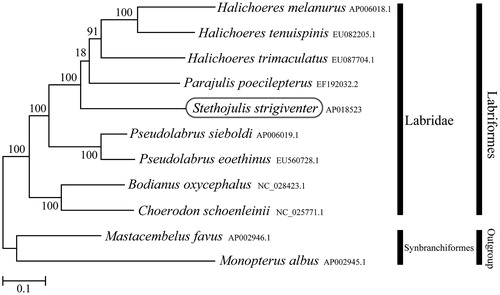
Phylogenetic trees were constructed by the maximum-likelihood method using MEGA 7.0 software (Kumar et al. Citation2016) for the newly sequenced genome and a further 10 complete mitochondrial genome sequences downloaded from the National Center for Biotechnology Information. We confirmed that S. strigiventer is clustered with genus Parajulis and Halichoeres, and formed monophyletic group with other Labridae species (genus Pseudolabrus, Bodianus and Choerodon) with high statistical support (). This mitochondrial genome provides important resources for addressing taxonomic issues and studying molecular evolution.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Barber PH, Bellwood DR. 2005. Biodiversity hotspots: evolutionary origins of biodiversity in wrasses (Halichoeres: Labridae) in the Indo-Pacific and new world tropics. Mol Phylogenet Evol. 35:235–253.
- Betancur-R R, Wiley EO, Arratia G, Acero A, Bailly N, Miya M, Lecointre G, Ortí G. 2017. Phylogenetic classification of bony fishes. BMC Evol Biol. 17:162.
- Bernardi G, Bucciarelli G, Costagliola D, Robertson DR, Heiser J. 2004. Evolution of coral reef fish Thalassoma spp. (Labridae). 1. Molecular phylogeny and biogeography. Mar Biol. 144:375–405.
- Cabanban A. 2010. Stethojulis strigiventer. The IUCN Red List of Threatened Species 2010: e.T187489A8548979. [accessed 2018 April 10]. http://dx.doi.org/10.2305/IUCN.UK.2010-4.RLTS.T187489A8548979.en.
- Clements KD, Alfaro ME, Fessler JL, Westneat MW. 2004. Relationships of the temperate Australasian labrid fish tribe Odacini (Perciformes; Teleostei). Mol Phylogenet Evol. 32:575–587.
- Hanel R, Westneat MW, Sturmbauer C. 2002. Phylogenetic relationships, evolution of broodcare behavior, and geographic speciation in the wrasse tribe Labrini. J Mol Evol. 55:776–789.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Maddison DR, Maddison WP. 2005. MacClade 4: analysis of phylogeny and character evolution. ver. 4.08. Sunderland (MA): Sinauer Associates.
- Rocha LA, Pinheiro HT, Gasparini JL. 2010. Description of Halichoeres rubrovirens, a new species of wrasse (Labridae: Perciformes) from the Trindade and Martin Vaz Island group, southeastern Brazil, with a preliminary mtDNA molecular phylogeny of new world Halichoeres. Zootaxa. 2422:22–30.
- Saccone C, De Giorgi C, Gissi C, Pesole G, Reyes A. 1999. Evolutionary genomics in Metazoa: the mitochondrial DNA as a model system. Gene. 238:195–209.