Abstract
Barnardia japonica is an ornamental bulb with important medicinal usage. The complete chloroplast genome of B. japonica was newly sequenced in this study. The total chloroplast genome size of B. japonica was 156,129 bp. In total, 131 genes were identified, including 85 protein-coding genes, 8 rRNA genes, and 38 tRNA genes. Eighteen genes are containing introns (clpP and ycf3 contained two introns) and 18 genes had two copies. The overall GC content of this genome was 37.7%. A further phylogenomic analysis of Asparagales, including 36 taxa, was conducted for the placement of genus Barnardia. The complete plastome of B. japonica will provide a valuable resource for further genetic conservation, phylogenomic, and evolution studies in the genus and family.
Asparagaceae, according to the APG III system (APG III Citation2009), in the order of Asparagales, is a family of monocotyledonous flowering plants, including 114 genera with a total of ∼2900 known species. Barnardia Lindley, is a small genus of bulbous flowering plants in the family Asparagaceae, subfamily Scilloidea. Only two species were included in this genus, Barnardia japonica and Barnardia numidica, which are mainly distributed in East Asia and Northwest Africa, Sounthwest Europe (Ali et al. Citation2012). B. japonica is heavily cultivated as an ornamental bulb showing attractive and sophisticated flowers. It also has been used for a wide range of medicinal applications to treat the rheumatism, cardiac, urinary infection, dermatological, and hemorrhoid disease (Chang Citation2017; Kayıran and Özkan Citation2017). Here, we assembled and characterized the first complete plastome of genus Barnardia. It will provide potential genetic resources for further evolutionary studies of the genus and other relatives in Asparaaceae.
Total DNA was extracted from fresh leaves of Barnardia japonica individual using DNA Plantzol Reagent (Invitrogen, Carlsbad). It is collected from Mingang, Wenzhou, Zhejiang, China (27°56'32.41"N, 120°31'34.06"E, Voucher No. WH1708010624, deposited at Zhejiang University). The plastome sequences were generated using Illumina HiSeq 2500 platform (Illumina Inc., San Diego, CA). In total, about 14.5 million high-quality clean reads (150 bp PE read length) were generated with adaptors trimmed. The CLC de novo assembler (CLC Bio, Aarhus, Denmark), BLAST, GeSeq (Tillich et al. Citation2017), and tRNAscan-SE v1.3.1 (Schattner et al. Citation2005) were used to align, assemble, and annotate the plastome.
The full length of Barnardia japonica chloroplast genome (GenBank Accession No. MH287351) was 156,129 bp and comprised of a large single copy region (LSC with 85,284 bp), a small single copy region (SSC with 18,429 bp), and two inverted repeat regions (IR with 26,208 bp). The overall GC content of the B. japonica cp genome was 37.7% and the GC content in the LSC, SSC, and IR regions are 35.8, 31.3, and 42.9%, respectively. A total of 131 genes were contained in the cp genome (85 protein-coding genes, 8 rRNA genes, and 38 tRNA genes. Eighteen genes had two copies, which included 6 PCG genes (ndhB, rpl2, rpl23, rps12, rps7, and ycf2), 8 tRNA genes (trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), and all 4 rRNA species (rrn4.5, rrn5, rrn16, and rrn23). Among the protein-coding genes, two genes (clpP and ycf3) contained two introns, and other ten genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, rps12, rps16) had one intron each.
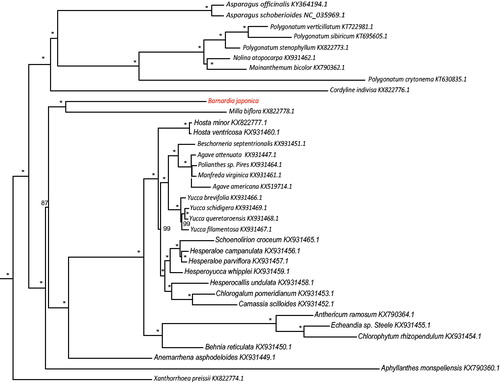
Thirty-six chloroplast genome of Asparagales were fully aligned with MAFFT v7.3 (Katoh and Standley Citation2013), and the maximum likelihood (ML) inference was performed using GTR + I + Γ model with 1000 bootstrap replicates with RAxMLv.8.2.1 (Stamatakis Citation2014) on the CIPRES cluster service (Miller et al. Citation2010). The result revealed that B. japonica was most closely related to members of genus Milla with the current sampling extent (). The newly characterized B. japonica complete chloroplast genome will provide essential data for further study on the phylogeny and evolution of the genus Barnardia and the family Asparagaceae.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Ali SS, Yu Y, Pfosser M, Wetschnig W. 2012. Inferences of biogeographical histories within subfamily Hyacinthoideae using S-DIVA and Bayesian binary MCMC analysis implemented in RASP (Reconstruct Ancestral State in Phylogenies). Ann Bot. 109:95–107.
- APG III. 2009. An update of the Angiosperm Phylogeny Group Classification for the orders and families of flowering plants: APG III. Bot J Linnean Soc. 161:105–121.
- Chang L. 2017. Asparagaceae. Identification and Control of Common Weeds. Vol. 3. Singapore: Springer. p. 835–859.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kayıran SD, Özkan EE. 2017. The ethnobotanical uses of Hyacinthaceae species growing in Turkey and a review of pharmacological activities. Indian J Tradit Knowledge. 16:243–250.
- Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE). IEEE, New Orleans, LA.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2014. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.