Abstract
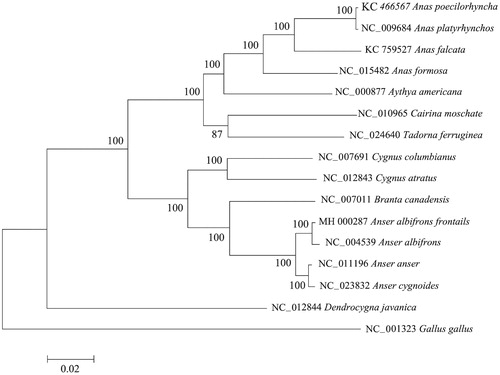
The mitochondrial genome of Anser albifrons frontails is 16740 bp in size, including 22 tRNAs, 2 rRNAs, 13 PCGs, and a CR. Except for tRNASer (AGY) and tRNALeu (CUN) without the dihydrouridine arm, all tRNAs could be folded into canonical cloverleaf structures. All PCGs start with ATG and the termination codon TAA is commonly observed. The CR is located between tRNAGlu and tRNAPhe. The result of the phylogenetic analysis suggests that Anser albifrons frontails is closely related to A. cygonoides and A. anser.
The Anser albifrons is a large water bird species, which belongs to the family Anatidae of order Anseriformes (IUCN Citation2017). Five subspecies were described in this species. The subspecies breeding in Northeast Siberia, W and N Alaska, wintering in USA, China and Japan, has been described as A. a. frontails (Dickinson Citation2003). A previous study found that A. a. frontalis was not regarded to merit subspecies status, it being synonymised with A. a. gambelli (Banks, Citation2011). The complete mitochondrial genome sequence is useful data for the phylogenetic study. In this study we determined the complete mtDNA sequence of the A. a. frontails with aiming to provide more useful molecular information for further studies of A. albifrons phylogeny.
We sequenced the complete mitogenome of A. a. frontails by PCR method. The tissue was sampled from an accidental death of individual from the Shengjin Lake National Nature Reserve (117°1′5.86″E,30°19′56.6″N), Anhui Province, in China on October 2013. It was kept in the institute of Biodiversity and wetland Ecology, Anhui University. The complete mtDNA sequence has been provided to GenBank with accession number MH000287.
The complete mitogenome of A. a. frontails is a 16740 bp circular molecular. The overall base composition of A, C, T, G are 30.14%, 32.09%, 22.61%, and 15.16%, respectively. A + T content is 52.75%, which is higher than C + G content (47.25%), similar to other Anseriformes (Liu et al. Citation2013). The mitogenome is comprised of 13 PCGs, 22 tRNAs, 2 rRNAs, and a CR; the gene order is identical to most of the other Anseriformes (Ely et al. Citation2005). Through the 13 PCGs, the longest one is ND5 (1817 bp), located between tRNALeu(CUN) and Cyt b, and the shortest is ATP8 (167 bp), which is between tRNALys and ATP6. All the start codons are typical ATG, expect COI, COII, and ND5 start with GTG, conventional stop codon TAA has been assigned to five of the PCGS, while the ND5 stop with AGA, ND1, and COI stop with AGG; ND2 and ND6 stops with TAG; COIII, and ND4 terminate with T-. The new mtDNA sequence contains a small subunit (12S rRNA) and a large subunit (16S rRNA) of rRNA, which is located between tRNAPhe and tRNALeu, separated by tRNAVal. The 12S rRNA and 16S rRNA are 988 bp and 1610 bp in length, respectively. All tRNA genes possess the typical clover leaf secondary structure except for tRNASer(AGY) and tRNALeu(CUN), which lacks a dihydroxyuridine (DHU) arm (Ely et al. Citation2005). The CR is located between tRNAGlu and tRNAPhe, and is 1177 bp long. The length of intergenic spacer sequences is 56 bp at 15 locations and the overlapping bases are 28 bp existing in 9 regions.
Phylogenetic tree was estimated using ML method, based on the complete mtDNA of 15 Anseriforme species, and corresponding Gallus gallus sequence was used as outgroup, sharing high node support values. From the phylogenetic tree, we can see that A. a. frontails first clustered with A. albifrons, and then together with other two species, including A. cygonoides and A. anser ().
Acknowledgements
The authors would like to thank Shenjin Lake National Nature Reserve for providing the specimen of A. a. frontails in this study.
Disclosure statement
The authors declare no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Banks RC. 2011. Taxonomy of greater White-fronted geese (Aves: Anatidae). Proc Biol Soc Washington. 124:226–233.
- Dickinson EC. 2003. The Howard and Moore Complete Checklist of the Birds of the World. 3rd ed. Princeton, NewJersey: Princeton University Press.
- Ely CR, Fox AD, Alisauskas RT, Andreev A, Bromley RG, Degtyarev AG, Ebbinge B, Gurtovaya EN, Kerbes R, Kondratyev AV, et al. 2005. Circumpolar variation in morphological characteristics of Greater White-Fronted Geese Anser albifrons. Bird Stud. 52:104–119.
- IUCN. 2017. IUCN Red List of Threatened Species. Version 2017-3. [accessed 2018 Apr 02]. <www.iucnredlist.org>.
- Liu G, Zhou LZ, Zhang LL, Luo ZJ, Xu WB. 2013. The complete mitochondrial genome of Bean goose (Anser fabalis) and implications for Anseriformes taxonomy. PLoS ONE. 8:e63334.