Abstract
The Old World climbing fern, Lygodium microphyllum, is a rapidly spreading environmental weed in Florida, United States. We reconstructed the complete chloroplast genome of L. microphyllum from Illumina whole-genome shotgun sequencing, and investigate the phylogenetic placement of this species within the Leptosporangiate ferns. The chloroplast genome is 158,891 bp and contains 87 protein-coding genes, four rRNA genes, and 27 tRNA genes. Thirty-three genes contained internal stop codons, a common feature in Leptosporangiate fern genomes. The L. microphyllum genome has been deposited in GenBank under accession number MG761729.
The Old World climbing fern, Lygodium microphyllum (Cav.) R. Br. (Pteridophyta: Lygodiaceae), is a fast-growing vining fern. It is native to Africa, Asia, and Australia, and has become naturalized in a number of regions, including south and central Florida (United States) (Pemberton and Ferriter Citation1998). It is especially damaging to the Everglades, where it forms thick mats across trees in the swamp, and contributes to fire hazard. Due to its significant ecological and economic impacts, it is the target of large-scale biological control programmes (see Goolsby et al. Citation2006; McCulloch et al. Citation2018). Here, we report the complete chloroplast sequence of this weed and investigate its phylogenetic placement within the Leptosporangiate ferns.
We assembled the complete chloroplast genome from a L. microphyllum specimen collected from Jacksonville, Florida (30.241421°N, –81.911946°W; voucher 2016JAX14, USDA-ARS Australian Biological Control Laboratory, Brisbane). DNA was extracted from leaf material using CTAB (Doyle Citation1987) followed by spin column purification (Ridley et al. Citation2016). A-sequencing library was constructed using the NebNext Ultra DNA kit. Sequencing was conducted at Novogene (Beijing, China) on the Illumina HiSeq 2500 platform, yielding 80 million paired-end 150-bp sequences. Sequences were iteratively mapped to the L. japonicum chloroplast genome (KC536645) in Geneious v11.0.3 (Kearse et al. Citation2012), and some de-novo assembly of chloroplast reads was performed to correct insertions and deletions. Gene annotations were made through comparison to L. japonicum, and manually checked and edited.
The complete chloroplast sequence of L. microphyllum is 158,891 bp, the second largest of 74 fern genomes sequenced to date. Gene content, G + C%, and gene order were similar to those of L. japonicum (Gao et al. Citation2013). The complete chloroplast genome contains 118 genes, including 87 protein-coding genes, four rRNA genes, and 27 tRNA genes. Six protein-coding genes, four rRNA genes, and five tRNA genes were duplicated (or partially duplicated) in the inverted repeat region. Thirty-three genes had internal stop codons and incorrect start codons, a common feature in the chloroplasts of Leptosporangiate ferns (see Wolf et al. Citation2004; Guo et al. Citation2015).
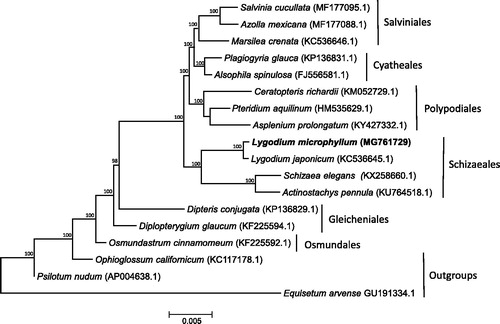
The phylogenetic placement of L. microphyllum within the Leptosporangiate ferns was assessed using Bayesian inference. Representative chloroplast genomes from six of the seven Leptosporangiate orders were downloaded from GenBank. In addition, three Eusporangiate ferns were downloaded and included as outgroups. Sequences were aligned using MAFFT (Katoh and Standley Citation2013) and non-coding regions were removed. A Bayesian phylogeny was constructed using MrBayes 3.2 (Huelsenbeck and Ronquist Citation2001) under the GTR + I + γ model of nucleotide substitution (). Four MCMC chains were run for 10,000,000 generations, with trees sampled every 1000 generations. Lygodium microphyllum is included in a well-supported clade with other Schizaeoid ferns (order: Schizaeales), with this clade sister to a clade containing the tree ferns (order: Cyatheales), Heterosporous ferns (order: Salviniales), and Polypod ferns (order: Polypodiales), consistent with previous phylogenies of this group (Pryer et al. Citation2004; Lu et al. Citation2015). This chloroplast genome provides a valuable resource for further resolving the evolutionary relationship among Lygodium species.
Acknowledgements
The authors thank Jessica Spencer (United States Army Corps of Engineers) for field assistance with sample collection, and Jeff Makinson (USDA-ARS ABCL) for lodging the voucher specimen. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the United States Department of Agriculture (USDA). USDA is an equal opportunity employer and provider.
Disclosure statement
The authors report no conflict of interest and are alone responsible for the content and writing of the paper.
Additional information
Funding
References
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Gao L, Wang B, Wang Z-W, Zhou Y, Su Y-J, Wang T. 2013. Plastome sequences of Lygodium japonicum and Marsilea crenata reveal the genome organization transformation from basal ferns to core leptosporangiates. Genome Biol Evol. 5:1403–1407.
- Goolsby JA, De Barro PJ, Makinson JR, Pemberton RW, Hartley DM, Frohlich DR. 2006. Matching the origin of an invasive weed for selection of a herbivore haplotype for a biological control programme. Mol Ecol. 15:287–297.
- Guo W, Grewe F, Mower JP. 2015. Variable frequency of plastid RNA editing among ferns and repeated loss of uridine-to-cytidine editing from vascular plants. Plos One. 10:e0117075
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17:754–755.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Lu JM, Zhang N, Du XY, Wen J, Li DZ. 2015. Chloroplast phylogenomics resolves key relationships in ferns. Jnl of Sytematics Evolution. 53:448–457.
- McCulloch GA, Makinson JR, Zonneveld R, Purcell MF, Raghu S, Walter GH. 2018. Assessment of genetic structuring in the Lygodium fern moths Austromusotima camptozonale and Neomusotima conspurcatalis in their native range: implications for biological control. Biol Control. 121:8–13.
- Pemberton RW, Ferriter AP. 1998. Old World climbing fern (Lygodium microphyllum), a dangerous invasive weed in Florida. Am Fern J. 88:165–175.
- Pryer KM, Schuettpelz E, Wolf PG, Schneider H, Smith AR, Cranfill R. 2004. Phylogeny and evolution of ferns (monilophytes) with a focus on the early leptosporangiate divergences. Am J Bot. 91:1582–1598.
- Ridley AW, Hereward JP, Daglish GJ, Raghu S, McCulloch GA, Walter GH. 2016. Flight of Rhyzopertha dominica (Coleoptera: Bostrichidae)-a Spatio-Temporal Analysis With Pheromone Trapping and Population Genetics . J Econ Entomol. 109:2561–2571.
- Wolf PG, Rowe CA, Hasebe M. 2004. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene. 339:89–97.