Abstract
Rhodiola rosea L. is used in herbal medicine in many countries for a long time. Here, its complete chloroplast genome was assembled and annotated. The genome is 151,348 bp long and comprises a pair of inverted repeat regions (IRs, 25,790 bp each), a large single-copy region (LSC, 82,716 bp), and a small single-copy region (SSC, 17,052 bp). It contained 113 gene species (79 protein coding, 29 tRNA, 4 rRNA, and 1 pseudogene), with 20 of them occurring in double copies. Introns were detected in 12 PCG and 5 tRNA species. The nucleotide composition is inhomogeneous (30.9% A, 19.2% C, 18.5% G, and 31.4% T) with an overall A + T content of 62.3%. Phylogenetic analysis indicated that Rhodiola rosea is sister to the remaining species of Rhodiola with maximum support in phylogeny.
Plants have been important footstones of a complex traditional medical system that has produced some of the most important drugs that still exist today (Gurib-Fakim Citation2006). Rhodiola rosea is a species of herbaceous perennials within the family Crassulaceae (Fu and Ohba Citation2001), whose roots and rhizomes are famous medicinal materials in many countries (Zhu et al. Citation2017). Here, we assembled its complete chloroplast (cp) genome and investigated its phylogenetic placement within Crassulaceae, to make use of R. rosea more efficiently (Famà et al. Citation2002; Mukherjee et al. Citation2016).
Using the CTAB method, total genomic DNA was isolated from silica-gel dried leaves of an individual of R. rosea from Mt. Xiaowutai (Hebei Province, China; N 39° 57′ 20″, E 115° 04′ 08″'). The voucher specimen was stored in the Peking University Herbarium (PEY) with accession number J. Q. Zhang et al. 120613-01. With R. ovatisepala as the starting reference, the clean reads were used to assemble the cp genome with the program MITObim v1.9 (Hahn et al. Citation2013) after 29.8 M of 150-bp raw paired reads were trimmed using CLC Genomics Workbench v8 (CLC Bio, Aarhus, Denmark). The genome was annotated with Geneious R10 (Biomatters Ltd., Auckland, New Zealand) using the cp genome of R. ovatisepala as the reference.
The cp genome of R. rosea is 151,348 bp long exhibiting a typical quadripartite structure. The large (LSC, 82,716 bp) and small (SSC, 17,052 bp) single-copy regions were separated by a pair of inverted repeat regions (IRs, 25,790 bp each). It contained 113 gene species (79 protein coding, 29 tRNA and 4 rRNA, and 1 pseudogene). About 20 gene species duplicated, including 9 PCG species (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf1, and ycf2), 7 tRNA species (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC), 4 rRNA species (4.5S, 5S, 16S, and 23S rRNA) and one pseudogene (ycf15). Of these 20 gene species, three species (ycf1, rps12, and rps19) partially within the IR regions, while the others completely within the IR regions. Fourteen gene species (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) contain one intron, three species (clpP, rps12 & ycf3) possess two introns. The nucleotide composition is asymmetric (30.9% A, 19.2% C, 18.5% G and 31.4% T) with an overall A + T content of 62.3%. The A + T content of the LSC (64.3%) and SSC (68.3%) regions are distinctly higher than those of the IR regions (57.1%).
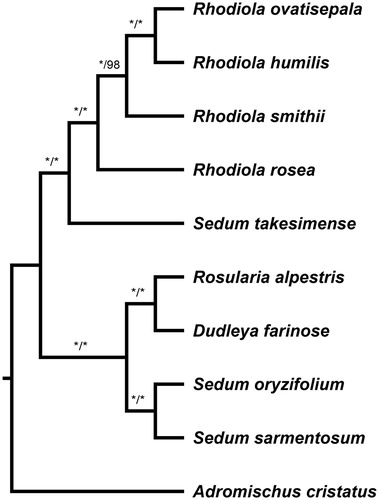
The most parsimony tree and Bayesian tree were conducted using PAUP* 4.0b10 (Swofford Citation2003) and MrBayes 3.2.1 (Ronquist and Huelsenbeck Citation2003), respectively, for 10 species. Three species of Sedum are obtained from GenBank (NC023085, NC026065, NC027837) and remaining species were from our own data. Outgroup was chosen as Adromischus cristatus. The ingroup can be subdivided into two clades with maximum support (). One clade comprised R. ovatisepala, R. humilis, R. smithii, R. rosea, and Sedum takesimense, and R. rosea is sister to the remaining species of Rhodiola; the other clade comprised Rosularia alpestris, Dudleya farinose, Sedum oryzifolium, and Sedum sarmentosum.
Acknowledgements
The authors thank Dr. Zengqiang Qian for technical help during the assembly of chloroplast genome.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Famà P, Jousson O, Zaninetti L, Meinesz A, Dini F, Di Giuseppe G, Iuseppe G, Millar AJK, Pawlowski J. 2002. Genetic polymorphism in Caulerpa taxifolia (Ulvophyceae) chloroplast DNA revealed by a PCR-based assay of the invasive Mediterranean strain. J Evol Biol. 15:618–624.
- Fu KT, Ohba H. 2001. Crassulaceae. In: Wu ZY, Raven PH, editors. Flora of China. Vol. 8. Beijing: Science Press.
- Gurib-Fakim A. 2006. Medicinal plants: traditions of yesterday and drugs of tomorrow. Mol Aspects Med. 27:1–93.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Mukherjee A, Williams D, Gitzendanner MA, Overholt WA, Cuda JP. 2016. Microsatellite and chloroplast DNA diversity of the invasive aquatic weed Hygrophila polysperma in native and invasive ranges. Aquat Bot. 129:55–61.
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19:1572–1574.
- Swofford D. 2003. PAUP: phylogenetic analysis using parsimony (*and other methods). Sunderland: Sinauer Associates.
- Zhu RW, Li YC, Zhong DL, Zhang JQ. 2017. Establishment of the most comprehensive ITS2 barcode database to date of the traditional medicinal plant Rhodiola (Crassulacaee). Sci Rep. 7:10051.