Abstract
We assembled the complete chloroplast genome of the Australian shrub Spyridium parvifolium var. parvifolium. The genome was 161,012 bp in length, with a pair of inverted repeats (IRs) of 26,515 bp, separated by a large single copy (LSC) region of 88,814 bp and a small single copy region (SCC) of 19,168 bp. The GC content was 36.9%. In total, 130 genes were annotated, including 86 protein coding genes, 36 tRNA genes and 8 rRNA genes. Phylogenetic analysis of 56 chloroplast genes placed this genome of S. parvifolium var. parvifolium within the family Rhamnaceae.
Spyridium Fenzl is a member of the cosmopolitan Rhamnaceae family (VicFlora Citation2016), which includes the economically important Chinese Date, Ziziphus jujuba Mill. (Liu et al. Citation2014). Spyridium parvifolium (tribe Pomaderreae) is a shrubby, widespread, and morphologically variable species from south-eastern Australia (Jessop et al. Citation1986; Curtis and Morris Citation1993; VicFlora Citation2016; PlantNET Citationn.d.). Several varieties of the species are sometimes recognized, and additional morphological variants have been identified (VicFlora Citation2016). Conflicting infraspecific taxonomies in different parts of Australia have implications for conservation management. In particular, two varieties (var. parvifolium and var. mole) are recognized in Tasmania, where both are listed as ‘Threatened’ under state legislation (Threatened Species Section Citation2016a, Citation2016b); in contrast, none of the varieties is currently recognized as distinct by the Australian Plant Census (CHAH Citation2016).
In this study, we report the complete chloroplast genome sequence of S. parvifolium var. parvifolium (GenBank accession MH234313). We generated this sequence to use as a reference in further chloroplast genome studies aimed at assessing phylogeography, genetic diversity, introgression, and infraspecific taxonomy of S. parvifolium.
Plant material was sampled from a population of var. parvifolium at Sisters Beach, Tasmania, Australia (40°54´15.0´´S 145°32´47.5´´E; Permit Number: TFL 15171; Voucher Specimen: MELUD155066a). Total DNA was extracted from leaves dried in silica gel using a modified CTAB protocol (Shepherd and McLay Citation2011), prepared for sequencing using the protocol of Schuster et al. (Citation2018), and sequenced on an Illumina NextSeq 550 (mid-output, 2 × 150 Paired End kit) at The Walter and Eliza Hall Institute of Medical Research (WEHI). The genome was assembled by mapping paired reads to the reference genome of Ziziphus jujuba (accession number KU351660). Contigs built in Spades 3.10.0 (Bankevich et al. Citation2012), CLC Genomics Workbench 10.0.1 and Geneious 10.2 (Kearse et al. Citation2012) were mapped to the consensus sequence for quality control. Annotations were transferred from the reference sequence, with reading frames reviewed and manually adjusted.
The complete chloroplast of S. parvifolium var. parvifolium was 161,012 bp in length. A pair of inverted repeats (IRs) of 26,515 bp were separated by a large single copy (LSC) region of 88,814 bp and a small single copy region (SCC) of 19,168 bp. The GC content of the chloroplast genome was 36.9%. In total 130 genes were annotated, including 86 protein coding genes, 36 tRNA genes, and 8 rRNA genes. One pseudogene was predicted (InfA) and two truncated repeats were recorded at IR boundaries (rps19 and ycf1). Annotations were identical between the reported genome (S. parvifolium var. parvifolium) and the reference (Ziziphus jujuba) except for one copy of the ycf1 gene which was annotated on the reported genome as protein coding while neither copy of the gene was annotated as protein coding on the reference genome.
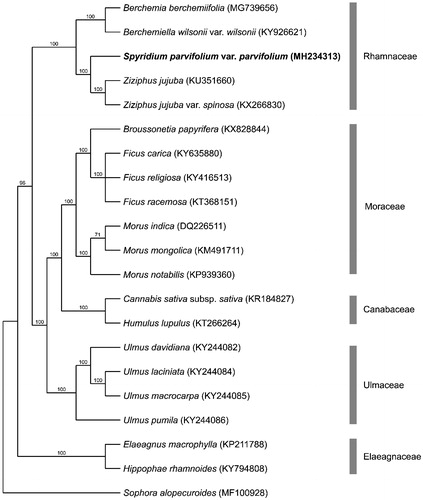
The phylogenetic tree presented in this study () builds from the results of Hauenschild et al. (Citation2016) and Cheon et al. (Citation2018). This tree shows S. parvifolium var. parvifolium within the Rhamnaceae clade and most closely related to Ziziphus jujuba (tribe Paliureae).
Figure 1. Bootstrap 50% majority rule consensus tree based on 56 protein coding chloroplast genes from 21 taxa including 20 species from the order Rosales and Sophora alopecuroides as the outgroup (CI = 0.8069 RI = 0.8744). Genes were aligned in MAFFT using default settings (Katoh et al. Citation2002). Sequences were analysed using maximum parsimony (MP) with PAUP 4.0a 161 using default settings (Swofford Citation2003). Bootstrap values are provided above branches. GenBank accessions are provided in brackets. Spyridium parvifolium var. parvifolium is highlighted in bold.
Acknowledgments
We acknowledge Mark Wapstra (ECOtas) for assisting with fieldwork. Jürgen Kellermann (State Herbarium of South Australia) for advice on Spyridium. Erin Batty and Todd McLay (The University of Melbourne) for support with DNA extractions. Stephen Wilcox (WEHI) for Illumina sequencing. Tanja Schuster (The University of Melbourne) for advice on preparation of this manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- CHAH. 2016. Autralian Plant Census (APC). [updated 2015 Jan 23; accessed 2016 Jul 25]. https://biodiversity.org.au/nsl/services/search?product=APC&tree.id=1133571&name=spyridium+parvifolium&inc._scientific=&inc.scientific=on&inc._cultivar=&max =100&display=apc&search=true.
- Cheon K-S, Kim K-A, Yoo K-O. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae). Mitochondrial DNA Part B. 3:133–134.
- Curtis WM, Morris DI. 1993. The student's flora of Tasmania. Hobart: St. David's Park Publishing.
- Hauenschild F, Matuszak S, Muellner-Riehl AN, Favre A. 2016. Phylogenetic relationships within the cosmopolitan buckthorn family (Rhamnaceae) support the resurrection of Sarcomphalus and the description of Pseudoziziphus gen. nov. Taxon. 65:47–64.
- Katoh K, Misawa K, Kuma Ki, Miyata T. 2002. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Jessop JP, Toelken HR, Black JM. 1986. Flora of South Australia. 4th ed. Adelaide: South Australian Government Printing Division.
- Liu M-J, Zhao J, Cai Q-L, Liu G-C, Wang J-R, Zhao Z-H, Liu P, Dai L, Yan G, Wang W-J, et al. 2014. The complex jujube genome provides insights into fruit tree biology. Nat Commun. 5:5315.
- PlantNET. n.d. The NSW Plant Information Network System. Royal Botanic Gardens and Domain Trust; [accessed 2018 Apr 30]. http://plantnet.rbgsyd.nsw.gov.au/cgi-bin/NSWfl.pl?page=nswfl&lvl=sp&name=Spyri-dium∼parvifolium.
- Schuster TM, Setaro SD, Tibbits JFG, Batty EL, Fowler RM, McLay TGB, Wilcox S, Ades PK, Bayly MJ. 2018. Chloroplast variation is incongruent with classification of the Australian bloodwood eucalypts (genus Corymbia, family Myrtaceae). PloS One. 13:e0195034.
- Shepherd LD, McLay TG. 2011. Two micro-scale protocols for the isolation of DNA from polysaccharide-rich plant tissue. J Plant Res. 124:311–314.
- Swofford DL. 2003. PAUP*:Phylogenetic analysis using parsimony, version 4.0a 161.
- Threatened Species Section. 2016a. Spyridium parvifolium var. molle (soft dustymiller): Species Management Profile for Tasmania's Threatened Species Link. Department of Primary Industries, Parks, Water and Environment, Tasmania; [accessed 2018 Apr 24]. Sequences were analysed using maximum parsimony (MP) with PAUP 4.0a 161 using default settings (Swofford 2003). http://www.threatenedspecieslink.tas.gov.au/Pages/Spyridium-parvifolium-var-molle.aspx.
- Threatened Species Section. 2016b. Spyridium parvifolium var. parvifolium (coast dustymiller): Species Management Profile for Tasmania's Threatened Species Link. Department of Primary Industries, Parks, Water and Environment, Tasmania; [accessed 2018 Apr 24]. http://www.threatenedspecieslink.tas.gov.au/Pages/Spyridium-parvifolium-var-parvifolium.aspx.
- VicFlora. 2016. Flora of Victoria. Royal Botanic Gardens Victoria; [accessed 2018 Apr 23]. https://vicflora.rbg.vic.gov.au.
