Abstract
Ulva is a green macroalga often causing a macroalgal bloom, ‘green tide’. Ulva ohnoi is a major species composing the green tide of the southern coastal regions of Japan. Here, we sequenced the complete mitochondrial and chloroplast genomes of the authentic strain of U. ohnoi. The mitochondrial and chloroplast genomes were of 65,326 bp and 103,313 bp, respectively, and the gene content was highly conserved in the Ulva species. The phylogenetic analyses using mitochondrial or chloroplast proteins represented the same topology with high supporting values. These results show that mitochondrial and chloroplast genomes can be used as reliable phylogenetic markers.
Ulva is a green macroalga in Ulvophyceae (Verbruggen et al. Citation2009). Species of Ulva are important for some applications, such as food stuffs (Hiraoka and Oka Citation2008; Holdt and Kraan Citation2011) and resource of biofuels (Bikker et al. Citation2016). In ecological aspects, some free-floating species of Ulva cause large-scale macroalgal bloom called ‘green tide’ (Fletcher Citation1996). Particularly, Ulva ohnoi is originally the major species composing the green tide in the southern Japanese coasts, such as Tosa bay and Hakata bay (Ohno Citation1988; Hiraoka, Ohno et al. Citation2004; Hiraoka, Shimada et al. Citation2004). In the past two decades, U. ohnoi bloom has been observed in Tokyo bay, which may have been introduced from tropical and subtropical waters (Yabe et al. Citation2009). Because it is difficult to distinguish Ulva species by morphological characters (Malta et al. Citation1999), molecular phylogenetic analyses were used to identify them; however, the complete organellar genomes of authentic strains of Ulva have not been reported.
Here, we sequenced the mitochondrial and chloroplast genomes of U. ohnoi KU-3321, which is an authentic strain of U. ohnoi established through parthenogenesis from subculture of gametes produced by the holotype specimen (strain KA43 in Hiraoka, Ohno et al. Citation2004). The strain was deposited in the Kobe University Macro-Algal Culture Collection (http://ku-macc.nbrp.jp/locale/change?lang=en). DNA was extracted from 2-week-old thalli. The paired-end libraries were sequenced on the MiSeq sequencing system (Illumina) with the MiSeq Reagent Kit v2 500 cycles. We acquired 9,172,559 read pairs, which were assembled using metaSPAdes v3.11.1 (Nurk et al. Citation2017). The 65,326-bp and 103,313-bp circular-mapping mitochondrial (AP018695) and chloroplast genomes (AP018696) were obtained, annotated by GeSeq (Tillich et al. Citation2017), and then manually curated. GC% of the mitochondrial and chloroplast genomes were 34.1% and 25.4%, respectively. The mitochondrial genome possessed 29 conserved protein-coding genes, 7 intronic ORFs, 2 hypothetical proteins, 2 rRNAs and 31 tRNAs. All the protein-coding genes were conserved among Ulva sp. UNA00071828 (Melton et al. Citation2015), U. pertusa (Liu et al. Citation2017), U. fasciata (Melton & Lopez-Bautista Citation2016), U. prolifera (Liu and Jun Pang Citation2015), U. linza (Zhou et al. Citation2016) and U. flexuosa (Cai, Wang, Jiang et al. Citation2017). U. ohnoi possessed trnS (GCA), but the gene was lacking in the other Ulva species. U. ohnoi possessed 8 introns; 1 intron each in atp1, cox2, nad3, rrs, and rrl, and 3 introns in cox1. The chloroplast genome possessed 71 conserved protein-coding genes, 7 intronic ORFs, 3 rRNAs and 26 tRNAs. All the genes except intronic ORFs were completely conserved among Ulva sp. UNA00071828 (Melton et al. Citation2015), U. fasciata (Melton and Lopez-Bautista Citation2017), U. prolifera (KX342867), U. linza (Wang et al. Citation2017) and U. flexuosa (Cai, Wang, Zhou et al. Citation2017).
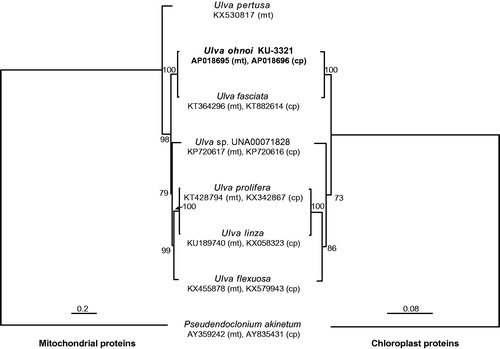
Phylogenetic analyses using 29 mitochondrial or 71 chloroplast proteins were performed (). Both trees represented the same topology with relatively high bootstrap support, and U. ohnoi was the sister of U. fasciata as the previous report (Hiraoka, Shimada et al. Citation2004). These results show that the mitochondrial and chloroplast genomes can be used as reliable phylogenetic markers.
Figure 1. Phylogenetic tree using mitochondrial (left) or chloroplast proteins (right). For mitochondrial tree (left), 8092 amino acids of 29 proteins were used. For chloroplast tree (right), 20,421 amino acids of 71 proteins were used. The amino acids were aligned using Mafft v7.164 (Katoh and Toh Citation2008), and highly divergent regions were manually trimmed using Mega 7 (Kumar et al. Citation2016). ML trees were inferred, and non-parametric bootstrapping analyses were performed 200 times using IQ-TREE 1.5.5 (Nguyen et al. Citation2015). The chloroplast genome of Ulva pertusa has been unavailable.
Acknowledgements
We thank Dr. Nobuyoshi Nakajima (National Institute for Environmental Studies) for sequencing the organellar genomes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bikker P, van Krimpen MM, van Wikselaar P, Houweling-Tan B, Scaccia N, van Hal JW, Huijgen WJJ, Cone JW, López-Contreras AM. 2016. Biorefinery of the green seaweed Ulva lactuca to produce animal feed, chemicals and biofuels. J Appl Phycol. 28:3511–3525.
- Cai C, Wang L, Jiang T, Zhou L, He P, Jiao B. 2017. The complete mitochondrial genomes of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta). Conserv Genet Resour. [accessed 2018 June 28]:[4 p.]. https://doi.org/10.1007/s12686-017-0838-6.
- Cai C, Wang L, Zhou L, He P, Jiao B. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLoS One. 12:e0184196
- Fletcher RL. 1996. The Occurrence of “Green Tides” – a Review. In: Schramm W, Nienhuis PH, editors. Mar Benthic Veg. Vol. 123. Berlin, Heidelberg: Springer; p. 7–43.
- Hiraoka M, Ohno M, Kawaguchi S, Yoshida G. 2004. Crossing test among floating Ulva thalli forming `green tide’ in Japan. Hydrobiologia. 512:239–245.
- Hiraoka M, Oka N. 2008. Tank cultivation of Ulva prolifera in deep seawater using a new “germling cluster” method. J Appl Phycol. 20:97–102.
- Hiraoka M, Shimada S, Uenosono M, Masuda M. 2004. A new green-tide-forming alga, Ulva ohnoi Hiraoka et Shimada sp. nov. (Ulvales, Ulvophyceae) from Japan. Phycological Res. 52:17–29.
- Holdt SL, Kraan S. 2011. Bioactive compounds in seaweed: functional food applications and legislation. J Appl Phycol. 23:543–597.
- Katoh K, Toh H. 2008. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinformatics. 9:286–298.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Liu F, Jun Pang S. 2015. The mitochondrial genome of the bloom-forming green alga Ulva prolifera. Mitochond DNA Part A. 5:1–2.
- Liu F, Melton JT, Bi Y. 2017. Mitochondrial genomes of the green macroalga Ulva pertusa (Ulvophyceae, Chlorophyta): novel insights into the evolution of mitogenomes in the Ulvophyceae. J Phycol. 53:1010–1019.
- Malta EJ, Kamermans P, Draisma S. 1999. Free-floating ulva in the southwest Netherlands: species or morphotypes? A morphological, molecular and ecological comparison. Eur J Phycol. 34:443–454.
- Melton JT, Leliaert F, Tronholm A, Lopez-Bautista JM. 2015. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta). PLoS One. 10:e0121020
- Melton JT, Lopez-Bautista JM. 2016. De novo assembly of the mitochondrial genome of Ulva fasciata Delile (Ulvophyceae, Chlorophyta), a distromatic blade-forming green macroalga. Mitochond DNA Part A. 27:3817–3819.
- Melton JT, Lopez-Bautista JM. 2017. The chloroplast genome of the marine green macroalga Ulva fasciata Delile (Ulvophyceae, Chlorophyta). Mitochond DNA Part A. 28:93–95.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 27:824–834.
- Ohno M. 1988. Seasonal changes of the growth of green algae, Ulva sp. in Tosa bay, southern Japan. Mar Fouling. 7:13–17 (in Japanese).
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.
- Verbruggen H, Ashworth M, LoDuca ST, Vlaeminck C, Cocquyt E, Sauvage T, Zechman FW, Littler DS, Littler MM, Leliaert F, et al. 2009. A multi-locus time-calibrated phylogeny of the siphonous green algae. Mol Phylogenet Evol. 50:642–653.
- Wang L, Cai C, Zhou L, He P, Jiao B. 2017. The complete chloroplast genome sequence of Ulva linza. Conservation Genet Resour. 9:463–466.
- Yabe T, Ishii Y, Amano Y, Koga T, Hayashi S, Nohara S, Tatsumoto H. 2009. Green tide formed by free-floating Ulva spp. at Yatsu tidal flat, Japan. Limnology. 10:239–245.
- Zhou L, Wang L, Zhang J, Cai C, He P. 2016. Complete mitochondrial genome of Ulva linza, one of the causal species of green macroalgal blooms in Yellow Sea, China. Mitochond DNA Part B. 1:31–33.
