Abstract
Pelecanus occidentalis, in the order Pelecaniformes, is one of the most abundant and widespread waterbird species in the coast of America. However, the phylogenetic relationships among Pelecaniformes, Suliformes, and Ciconiiformes remain unresolved, particularly in Pelecanidae and Ciconiidae. In this study, we first sequenced and described the complete mitochondrial genome and phylogeny of P. occidentalis. The whole genome of P. occidentalis was 17,315 bp in length, and contained 13 protein-coding genes, 21 transfer RNA genes, two ribosome RNA genes, and one non-coding control region. The overall base composition of the mitochondrial DNA was 30.1% for A, 23.7% for T, 31.5% for C, and 14.6% for G, with a GC content of 46.1%. A phylogenetic tree confirmed that P. occidentalis (Pelecaniformes) was sister to C. boyciana (Ciconiiformes), and Ardeidae and Threskiornithidae were both monophyletic group. This information will be useful in the current understanding of the phylogeny and evolution of Pelecaniformes.
Pelecanus occidentalis, in the order Pelecaniformes, is one of the most abundant and widespread waterbird species in the coast of America. This species is listed as of Least Concern (LC) on the IUCN Red List of Threatened Species (IUCN Citation2017). Despite this, genetic information of P. occidentalis is quite limited, which lead to confused and contentious phylogenetic relationships among Pelecaniformes, Suliformes, and Ciconiiformes (Zhang et al. Citation2012). Normally, the Ciconiiformes is restricted to included only the family Ciconiidae, and the Pelecaniformes consists of Pelecanidae, Ardeidae, Threskiornithidae, Balaenicipitidae, and Scopidae (Clements et al. Citation2017) (based on the recommendation of the North American Classification Committee). However, this taxonomy has been questioned by Gibb (Gibb et al. Citation2013) and Zhang (Zhang et al. Citation2012), particularly in Pelecanidae and Ciconiidae. Therefore, we sequenced the complete mitochondrial genome of P. occidentalis to enhance our understanding of the phylogeny and evolution of Pelecaniformes.
The specimen was collected from southeastern United States and stored at Hunan Engineering Research Center for Internet of Animals, China. The total mitochondrial DNA was extracted from the muscle tissue using a DNeasy Plant Mini Kit (Qiagen, Valencia, CA), and sequenced by using the Illumina Miseq Platform (Illumina, San Diego, CA). Adapters and low-quality reads were removed using the NGS QC Toolkit (Patel and Jain Citation2012). The genome was annotated using the MITOS WEB Server (Bernt et al. Citation2013). Each of the annotated peotein-coding genes (PCGs) were compared with published sequences of other vertebrate species using DOGMA (Wyman et al. Citation2004). Drawing a circular map of the mitochondria referred to Zhang’s method (Zhang et al. Citation2017) by using OGDRAW (Lohse et al. Citation2007). Phylogenetic tree of the relationships among Pelecaniformes and its related orders were presented using 13 shared PCGs among 18 species by Neighbour-joining (NJ) analyses using MEGA 7.0 software with 1000 bootstrap replication (Kumar et al. Citation2016). The complete mitochondrial genome of P. occidentalis was submitted to the NCBI database under the accession number MH041272.
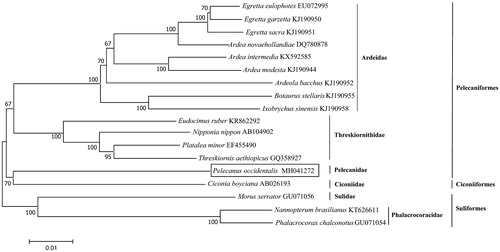
The complete mitochondrial genome of P. occidentalis was 17,315 bp in length. A total of 36 mitochondrial genes were identified, including 13 protein-coding genes (PCGs), 21 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one non-coding control region (D-loop). Among these genes, ND6 and eight tRNAs (trnGln, trnAla, trnAsn, trnCys, trnTyr, trnSer, trnPro, and trnGlu) were located on the light strand (L-strand), while all of the remaining genes were located on the heavy strand (H-strand). The overall base composition of P. occidentalis mitogenome was 30.1% for A, 23.7% for T, 31.5% for C, and 14.6% for G, with a GC content of 46.1%. The reconstructed phylogenetic tree supported the placement of P. occidentalis in Pelecaniformes (Ardeidae, Pelecanidae, Phalacrocoracidae, and Threskiornithidae), Suliformes (Sulidae), and Ciconiiformes (Ciconiidae) clade (). All of the nodes were inferred with strong support by the NJ analysis. Our results confirmed that P. occidentalis (Pelecaniformes) was sister to C. boyciana (Ciconiiformes), and Ardeidae and Threskiornithidae were both monophyletic group. In all, the mitochondrial genome reported here would be useful in the current understanding of the phylogeny and evolution of Pelecaniformes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313.
- Clements JF, Schulenberg TS, Iliff MJ, Roberson D, Fredericks TA, Sullivan BL, Wood CL. 2017. The eBird/Clements checklist of birds of the world: v2016. http://www.birds.cornell.edu/clementschecklist/download/
- Gibb GC, Kennedy M, Penny D. 2013. Beyond phylogeny: pelecaniform and ciconiiform birds, and long-term niche stability. Mol Phylogenet Evol. 68:229–238.
- IUCN. 2017. The IUCN Red List of Threatened Species. Version 2017-3. [Downloaded on 11 May 2018]. www.iucnredlist.org
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52:267–274.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. Plos One. 7:e30619.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Zhang LQ, Wang L, Gowda V, Wang M, Li XF, Kan XZ. 2012. The mitochondrial genome of the Cinnamon Bittern, Ixobrychus cinnamomeus (Pelecaniformes: Ardeidae): sequence, structure and phylogenetic analysis. Mol Biol Rep. 39:8315–8326.
- Zhang W, Zhao YL, Yang GY, Tang YC, Xu ZG. 2017. Characterization of the complete chloroplast genome sequence of Camellia oleifera in Hainan, China. Mitochondr DNA B. 2:843–844.