Abstract
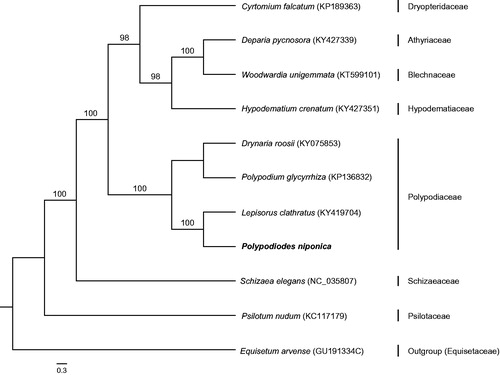
The complete chloroplast genome of Polypodiodes niponica has been determined. It is a double stranded circular DNA of 152,551 bp length, containing two inverted repeat (IR, 24,643 bp each) regions, separated by large single-copy (LSC, 81,506 bp) and small single-copy (SSC, 21,759 bp) regions, respectively. The genome contains 131 genes, including 88 protein-coding genes, 35 tRNA genes, and eight rRNA genes. The total GC content of the chloroplast genome is 42.4%. ML phylogenetic analysis revealed that P. niponica was most closely related to Lepisorus clathratus.
Polypodiodes niponica, commonly known as Polypodium niponicum Mett. or Marginaria niponica Nakai, is a polypodiaceous fern. It is epiphytic on tree trunks or on rocks with elevation between 300–1800 m. Its rhizome is gray-green, long creeping, and sparsely covered with whitish bloom and scales (Zhang et al. Citation2013). The plant is extensively distributed in Japan and China such as Anhui, Fujian, Gansu, Guangdong etc. In China, it is not only an edible fern (Liu et al. Citation2012), but also an important medical plant known as “Shican” which is used in clearing heat and promoting diuresis (Wang et al. Citation2009). As a key genus in Polypodioideae, Polypodiodes plays an important role in exploring the phylogenetic relationships among Polypodiastrum, Schellolepis, and Goniophlebium, which occurs a great controversy (Schneider et al. Citation2004; Lu and Li Citation2006). Hence, the acquirement of whole chloroplast (cp) genome of Polypodiodes niponica will contribute to further development of the medicinal resources and survey phylogeny of the family Polypodioideae.
Total genomic DNA was isolated from fresh leaves sampled from the South China Botanical Garden (23°11′3.56″N, 113°21′43.28″E) using Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China). The specimen is stored in Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 20161013). On an average 300 bp DNA fragment was obtained using Covaris M220 (Covaris Inc., MS, USA), which was further used to construct genomic library. Sequencing was performed on Illumina Hiseq 2500. Trimmomatic v0.32 was employed to filter and trim low-quality reads (Bolger et al. Citation2014). After visual evaluation by FastQC v0.10.0 (Andrews Citation2010), we assembled high-quality clean reads into the chloroplast genome using Velvet v1.2.07 (Zerbino and Birney Citation2008) and annotated with DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005). Finally, BLASTX and BLASTN were used to validate the location of encoding genes and RNAs. The complete chloroplast genome sequences of 11 ferns (Equisetum arvense as outgroup) were aligned by MAFFT (Katoh et al. Citation2002) and phylogenetic tree was inferred based on the maximum likelihood (ML) analysis using RAxML v.8.0 with 1000 bootstrap replicates (Stamatakis Citation2014).
The complete chloroplast genome of P. niponica is a double stranded circular DNA with 152, 551 bp length, containing two inverted repeat (IR) regions of 24,643 bp each, separated by large single-copy (LSC) and small single-copy (SSC) regions of 81,506 bp and 21,759 bp, respectively (GenBank accession number: MH319944). A total of 131 functional genes are successfully annotated, including 88 protein-coding genes (PCGs), 35 tRNA genes and eight rRNA genes. Among them, 14 are duplicated in the IR regions, including four PCGs, six tRNA genes and four rRNA genes. The GC content of the whole chloroplast genome is 42.4%, which is higher than that of LSC and SSC (41.5% and 39.0%, correspondingly). The IR region possesses the highest GC content (45.3%). Furthermore, there are 18 intron-containing genes, almost all of which are single-intron genes except for ycf3, clpP, and rps12 with two introns. ML tree indicated that P. niponica was most closely related to Lepisorus clathratus with high support value (). The chloroplast genome of P. niponica provides valuable molecular data for development of the medical fern and phylogeny of ferns.
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge (UK): the Babraham Institute; [accessed 2017 Jul 15]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Lu SG, Li CX. 2006. Phylogenetic position of the monotypic genus Metapolypodium Ching endemic to Asia: evidence from chloroplast DNA sequences of rbcL gene and rps4-trnS region. Acta Phytotaxon Sin. 44:494–502.
- Liu YJ, Wujisguleng W, Long CL. 2012. Food uses of ferns in China: a review. Acta Soc Bot Pol. 81:263–270.
- Schneider H, Smith AR, Cranfill R, Hildebrand TJ, Haufler CH, Ranker TA. 2004. Unraveling the phylogeny of polygrammoid ferns (Polypodiaceae and Grammitidaceae): exploring aspects of the diversification of epiphytic plants. Mol Phylogenet Evol. 31:1041–1063.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Wang YL, Gao H, Wang MH, Feng XL, Zhang L. 2009. Determination of total flavonoids in Polypodiodes niponica. Chin Wild Plant Resour. 28:62–66.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
- Zhang XC, Lu SG, Lin YX, Qi XP, Moore S, Xing FW, Wang FG, Hovenkamp PH, Gilbert MG, Nooteboom HP, et al. 2013. Polypodiaceae. In: Wu ZY, Raven PH, Hong DY, eds., Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 758–850.