Abstract
Histiopteris incisa is a core member to address the issue of relationship between Histiopteris and Pteris. Its complete chloroplast genome was generated by de novo assembly using Illumina sequencing. The complete chloroplast genome is a quadripartite structure of 16,0567 bp in length, with two inverted repeat regions (IRs) of 27,119 bp each, a large single-copy (LSC) region of 84,897 bp and a small single-copy (SSC) region of 21,432 bp. A total of 132 genes were predicted, involving in 88 protein-coding genes, eight rRNA genes, 35 tRNA genes, and one pseudogene. ML phylogenetic analysis showed that H. incisa was closely related to Pteridium aquilinum subsp. aquilinum.
Histiopteris incisa is only one accepted species of Histiopteris in the family Dennstaedtiaceae (Yan et al. Citation2013). The fern is up to 2 m tall with long-creeping rhizome covered with chestnut-brown scales (Yan et al. Citation2013). Known as bat's wing fern or water fern, its fronds are soft and green and have deeply lobed segments. The plant is usually found in the moist areas, where it may form extensive colonies. It is widespread across Bhutan, NE India, S Japan, pantropical areas, islands near Antarctica, and Madagascar (Yan et al. Citation2013). Histiopteris incisa is a core member to explore the phylogenetic relationships between Histiopteris and Pteris due to their morphological similarity (Yan et al. Citation2013). Hence, acquirement of its whole chloroplast (cp) genome sequence will lay solid foundation to address this issue.
Sample of H. incisa was provided by South China Botanical Garden, Chinese Academy of Sciences (23°19’28.21’’N, 113°37’47.46’’E). The specimen is stored in Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 20161012). Total genomic DNA was isolated by Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China). Average 300 bp Illumina paired-end genomic library was prepared and sequenced on an Illumina Hiseq 2500 platform (Illumina Inc., San Diego, CA). After trimming the sequences using Trimmomatic v0.32 (Bolger et al. Citation2014), we obtained high quality clean reads and assessed their quality by FastQC v0.10.0 (Andrews Citation2010). These clean reads were further de novo assembled into contigs by Velvet v1.2.07 (Zerbino and Birney Citation2008). DOGMA (Wyman et al. Citation2004) and tRNAscan-SE (Schattner et al. Citation2005) were used to conduct the annotation.
The complete chloroplast genome of H. incisa (GenBank accession number: MH319942) is a quadripartite structure of 16,0567 bp in length, with two inverted repeat regions (IRs) of 27,119 bp each, a large single-copy (LSC) region of 84,897 bp and a small single-copy (SSC) region of 21,432 bp. It harbors 132 genes, including 88 protein-coding genes, eight rRNA genes, 35 tRNA genes, and one pseudogene. A total of 18 genes have introns. Among them, ndhB, rps16, atpF, rpoC1, petB, petD, ndhA, rpl16, and rpl2 have a single intron, whereas ycf3, clpP, and rps12 contain two introns. Fourteen genes occur as two copies, including four protein- coding genes (rps12, rps7, psbA, and ycf2), six tRNA genes (trnN-GUU, trnH-GUG, trnI-GAU, trnA-UGC, trnT-UGU, and trnR-ACG), and four rRNA genes (rrn4.5, rrn5, rrn16, and rrn23). The total GC content was 43.0% and the corresponding values in LSC, IRs, and SSC are 41.2%, 46.5%, and 39.1%, respectively.
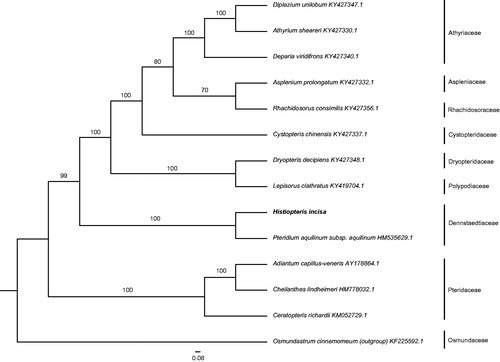
Phylogenetic analysis was carried out using the complete genome sequence of 14 ferns and Osmundastrum cinnamomeum as outgroup by Maximum likelihood (ML) method. After sequence alignment by MAFFT (Katoh et al. Citation2002), phylogenetic tree was constructed using RAxML (Stamatakis Citation2014) with 1000 bootstrap replicates. The results revealed that H. incisa was closely related to Pteridium aquilinum subsp. aquilinum (). The complete chloroplast genome of H. incisa provides powerful data to further elucidate phylogenetic relationships of dennstaedtioid ferns.
Disclosure statement
The authors declare no conflict of interests. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data; [accessed 2017 Sep 2]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.
- Yan YH, Qi XP, Liao WB, Xing FW, Ding MY, Wang FG, Zhang XC, Wu ZH, Serizawa S, Prado J, et al. 2013. Dennstaedtiaceae. In: Wu ZY, Raven PH, Hong DY, eds., Flora of China. Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 147–168.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.