Abstract
The complete chloroplast genome of Tetradoxa omeiensis, which is endemic and endangered to the Hengduan Mountains, was sequenced and characterized in this study. The chloroplast genome is 157,502 bp in length, and it is consisted of four distinct regions: a large single-copy region (LSC, 86,526 bp), a small single-copy region (SSC, 18,682 bp) and a pair of inverted repeat regions (IRs, 26,147 bp). A total of 134 genes were annotated, including 86 protein-coding genes (80 PCG species), 40 tRNA genes (33 tRNA species) and 8 ribosomal RNA genes (4 rRNA species). We also reconstructed a comprehensive phylogenetic relationship of the family Adoxaceae, which demonstrated that the three Adoxaceae species clustered into two well-supported clades, and T. omeiensis is most closely related to the Adoxa moschatellina.
Tetradoxa omeiensis, a monotypic plant of the genus Tetradoxa (Adoxaceae), is endemic to Hengduan Mountains, where is the southeastern margin of the Qinghai-Tibet Plateau of China (Wu Citation1981). This species is listed as “Endangered” in the latest IUCN Red List of Threatened Species (http://www.iucnredlist.org/search) and is also an endangered species in China (Wang and Xie Citation2004). In recent years, the related researches about T. omeiensis is rare and conflicting (Mao et al. Citation2005). The accurate phylogenetic relationship for T. omeiensis need to be better studied as a conservation target for further protection and recovery. Herein, we reported the complete chloroplast genome and the gene annotation of T. omeiensis. The annotated chloroplast genome has been submitted to GenBank under the accession number of KX258653.
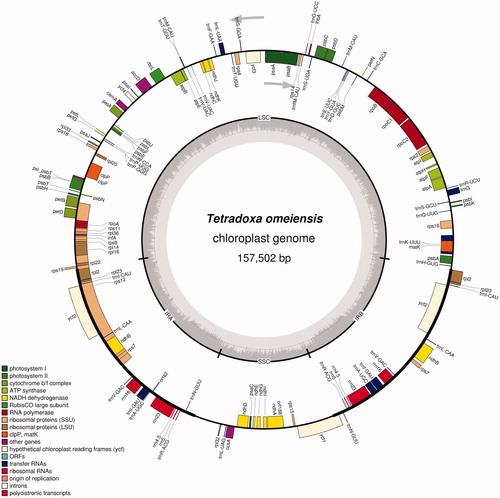
Fresh leaves of a single individual were collected from Erlangshan (Sichuan, China; coordinates: 102°18'9" E, 29°50'41" N). Voucher specimen of the species was deposited in the Key Laboratory of Bio-resource and Eco-environment of Ministry of Education (Sichuan, China). The leaves were used to extract the total genomic DNA with a modified CTAB method (Doyle and Doyle Citation1987). The whole-genome sequencing was carried out on the Hiseq2500 System (Illumina, USA). In all, about 2 G high-quality base pairs of sequencing data were obtained and used for the genome assembly using Velvet version 1.2.04 (Zerbino and Birney Citation2008). Then, the resulting contigs were aligned to the reference Sinadoxa corydalifolia chloroplast genome (KX258651; Wang et al. Citation2016) with Samtools (Li et al. Citation2009) and further processed into a complete genome by Genious version 8.0.5 (Kearse et al. Citation2012). After filling gaps with GapCloser (Luo et al. Citation2012), a 157,502-bp chloroplast genome of T. omeiensis was obtained. The annotation was performed using Plann (Huang and Cronk Citation2015). Then, we corrected the annotation with Sequin software (NCBI website). Finally, a physical map was drawn using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/; Lohse et al. Citation2013; ).
The complete chloroplast genome of T. omeiensis has a total length of 157,502 bp with a typical quadripartite structure, consisting of a large single-copy (LSC) region of 86,526 bp, two inverted repeat (IR) regions of 26,147 bp and a small single-copy (SSC) region of 18,682 bp. A total of 134 genes were annotated, including 86 protein-coding genes (80 PCG species), 40 tRNA genes (33 tRNA species) and 8 ribosomal RNA genes (4 rRNA species). The most of gene species occur as a single copy, while 17 gene species occur in double copies, including all rRNA species, 7 tRNA species and 6 PCG species.
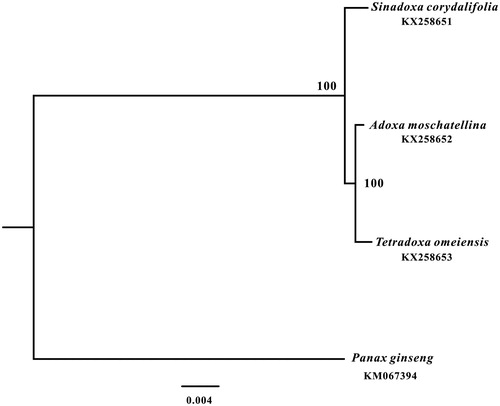
We also reconstructed a comprehensive phylogenetic tree of the family Adoxaceae, which consisted of only three genus and each genus contained only one species (Wu Citation1981), based on their complete chloroplast genome sequences. We aligned the sequences with MAFFT version 7.221 (Katoh and Standley Citation2013). And the maximum likelihood (ML) tree with 1000 bootstrap replicates was performed with RAxML version 8.1.24 (Stamatakis Citation2014). The ML phylogenetic tree strongly showed that Adoxaceae species clustered into two well-supported clades, and T. omeiensis was closely related to Adoxa moschatellina ().
Above all, this report provides essential data for population, phylogenetic, and other studies of T. omeiensis, as well as Adoxaceae. A good knowledge of its genomic information can provide insights for development, utilization and evolutionary histories.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3:1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, and 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25:2078–2079.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:W575–W581.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, et al. 2012. Soapdenovo2: an empirically improved memory-efficient short-read de novo, assembler. GigaScience. 1:1–6.
- Mao KS, Yao XL, Huang ZH. 2005. Molecular Phylogeny and Speciation of Adoxaceae s.s. Acta Bot Yunnanica. 27:620–628.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang S, Xie Y. 2004. China Species Red List. Vol. 1. Beijing: Red List.
- Wang YL, Guo XY, Hao GQ, Wang TJ, Wang K. 2016. The complete chloroplast genome of Sinadoxa corydalifolia (Adoxaceae). Conservation Genet Resour. 8:303–305.
- Wu CY. 1981. Another new genus of Adoxaceae, with special references on the infrafamiliar evolution and the systematic position of the family. Acta Bot Yunnan. 3:383–388.
- Zerbino DR, Birney E. 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.