Abstract
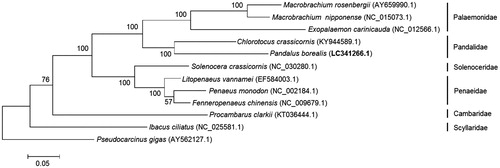
Pandalus borealis is an important indicator species to study the state of the Arctic ecosystem. The mitochondrial genome of P. borealis is 15,956 bp in length and encodes 13 protein-coding genes. The phylogenetic tree of eleven shrimps revealed that P. borealis belonged to Pandalidae family and was closely related to C. crassicornis. This mitogenome will be of significance to study the Arctic ecosystem state and perform the resource protection of this species.
Pandalus borealis is the most abundant cold-water shrimp around the Arctic (Jiao et al. Citation2015). Besides the commercial value, it is an important indicator to study the state of the Arctic ecosystem (Anderson Citation2000). Temperature, acidification, salinity, and fishing impact the global catch of this species (Bergstrom Citation2000; Bechmann et al. Citation2011). The catch has decreased from 446,909 tons (2004) to 261,435 tons (2014; http://www.fao.org/fishery/species/3425/en), suggesting that the climatic change and fishing might influence the genetic resource of P. borealis. Herein, we characterized the mitochondrial genome of P. borealis (GenBank Accession: LC341266.1).
The captive specimen and the extracted DNA samples were deposited in our laboratory (Centre for Applied Aquatic Genomics, Chinese Academy of Fishery Sciences). The genomic DNA was used to construct one paired-end library. The library was sequenced on the Illumina platform with 150 PE mode. After trimmed using SolexaQA (Cox et al. Citation2010), the cleaned reads (NCBI Sequence Read Archive Accession: SRX3443045) were assembled to contigs using SOAPdenovo (Luo et al. Citation2012). The contigs homologous to the mitochondria genomes of close shrimp were scaffolded into one long sequence using SSPACE (Boetzer et al. Citation2011). The gaps were filled with Gapcloser (Luo et al. Citation2012).
The mitogenome is 15,956 bp in length and exhibits a high A + T content of 65.57%. Thirteen protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes are identified using MITOS (Bernt et al. Citation2013). Six PCGs (COX2, ATP6, COX3, NAD4, NAD4L, and COB) initiate with ATG and four PCGs (COX1, ATP8, NAD5, and NAD6) with ATC. The rest PCGs use ATA and ATT as the start codons. Eleven PCGs use TAA as the stop codon and the rest two PCGs (COX2 and COB) end with the incomplete stop codon T (Boore Citation1999).
A phylogenetic tree was constructed using Maximum likelihood (ML) analysis in MEGA package (Kumar et al. Citation2016) under the Jones-Taylor-Thornton model with 1000 bootstrap replicates. This tree was based on 13 mitochondrial PCGs of eleven shrimps and Pseudocarcinus gigas (as the outgroup). P. borealis was grouped with Chlorotocus crassicornis, both of which belong to Pandalidae family (). Because of little genetic information for this family, this mitogenome will provide basic genetic data for phylogeny study. Furthermore, the result is of significance to dynamically study the Arctic ecosystem state.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Anderson P. 2000. Pandalid shrimp as indicators of ecosystem regime shift. J Northwest Atlantic Fishery Sci. 27:1–18.
- Bechmann RK, Taban IC, Westerlund S, Godal BF, Arnberg M, Vingen S, Ingvarsdottir A, Baussant T. 2011. Effects of ocean acidification on early life stages of shrimp (Pandalus borealis) and mussel (Mytilus edulis). J Toxicol Environ Health A. 74:424–438.
- Bergstrom B. 2000. The biology of Pandalus. Adv Marine Biol. 38:55–245.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. 2011. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 27:578–579.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27:1767–1780.
- Cox MP, Peterson DA, Biggs PJ. 2010. SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform. 11:485.
- Jiao G, Hui JP, Burton IW, Thibault MH, Pelletier C, Boudreau J, Tchoukanova N, Subramanian B, Djaoued Y, Ewart S, et al. 2015. Characterization of shrimp oil from Pandalus borealis by high performance liquid chromatography and high resolution mass spectrometry. Marine Drugs. 13:3849–3876.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 1:18.