Abstract
Macrocystis integrifolia, one species of brown alga, is one of the largest and most complex and dynamic ecosystems in the ocean, which can be also used as plant fertilizer. In the present study, the complete mitochondrial genome of M. integrifolia was reconstructed from whole genome Illumina sequencing survey data. The size of M. integrifolia mitochondrial DNA is 37,366 bp, including 3 rRNAs, 25 tRNAs, 35 proteins and 2 ORFs. The phylogenetic tree, which was established based on mitochondrial genomes of brown algae, indicated that M. integrifolia is at an earlier stage of differentiation in Laminariaceae.
In the northern hemisphere, the brown algae include many types of seaweed that are important members of the Marine ecosystem. Because they not only create habitats, but also provide food for other organisms (Hoek et al. Citation1995). Macrocystis integrifolia is one species of kelp (large brown algae) in the genus Macrocystis, which contains the largest of all the Phaeophyceae or brown algae. It is typically found on intertidal rocks or shallow subtidal rocks of British Columbia, Mexico, Peru and Northern Chile, which grows to about 6 m long (Chihara Citation1977; Druehl Citation1978; Graham et al. Citation2007). M. integrifolia is considered the main part of the temperate forest of seaweed, which is one of the largest and most complex and dynamic ecosystems in the ocean for their biological significance. Previous studies showed that the M. integrifolia was used as plant fertilizer, increasing the yield of soybean 24%, and chemical studies showed that there was a hormone-like substance (Temple et al. Citation1989).
The mitochondrion is responsible for respiration of biology and can be used as molecular marks which provide useful and effective for species identification, phyletic evolution, and genetic structure (Galtier et al. Citation2009; Cai et al. Citation2017).
In the present study, we assemble the complete mitochondrial genome of M. integrifolia (GenBank accession MH411105) from previously genome survey data. Specimens were collected at Perharidy, France (48°72′N,4°01′W). Both DNA and voucher specimen were resided at the Gene Denovo (Guangzhou, China). The complete mitogenome sequence is 37,366 bp in length, with a nucleotide composition (29.7%A, 19.2%G, 12.8%C, and 38.4%T). Moreover, a total of 65 genes were identified, including 35 protein-coding genes, 25 transfer tRNA genes, 3 ribosomal RNAs (5s rRNA, 16s rRNA, 23s rRNA), and 2 ORFs (ORF130, ORF377). Similar to the known brown algae mitochondrial genome, the M. integrifolia genome contains 35 protein-coding genes (rps2-4, rps7-8, rps10-14, rps19, atp6, atp8, atp9, cox1-3, nad1-7, nad9, nad11, nad4L, rpl2, rpl5, rpl6, rpl14, rpl16, rpl31, tatC, and cob). Most protein-coding genes and ORFs have a conserved, typical ATG codon as a start signal (34/37), while the gene of rps10 and cox2-3 used TTG as the initial codon. In addition, the most common stop codon is TAA (29 of 37 genes), whereas TAG and TGA are used as translation termination codon of 4 (nad4, rpl5, cox3, rps12) and 4 genes (nad3, rps7, rps8, tatC).
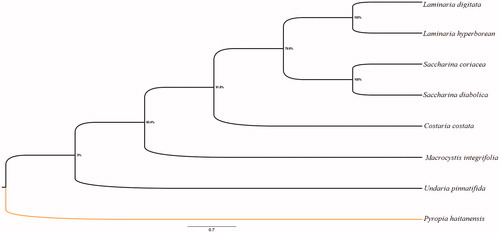
To further validate the newly determined sequence, the program MEGA6 was used to reconstruct the phylogenetic tree based on the maximum likelihood (ML). Phylogenetic analysis shows that all species are divided into two branches, the Pyropia haitanensis is alone, and the remaining species are clustered into a branch. In addition, the results also show that Laminaria digitata and Laminaria hyperborean clustered together, the Saccharina Diabolica is closed to the Saccharina coriacea, and M. integrifolia is at an earlier differentiation position ().
Figure 1. Phylogenetic trees (1000 bootstrap replicates) derived from maximum likelihood (ML) based on 13 mitochondrial PCGs.
This complete mtDNA genome can be subsequently used for population genomic studies of M. integrifolia and such information would provide a platform for the study of this species in biology.
Disclosure statement
The authors declare that they have no competing interests. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Cai C, Wang L, Jiang T, Zhou L, He P, Jiao B. 2017. The complete mitochondrial genomes of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta). Conserv Genet Resour. 10:415–418.
- Chihara M. 1977. Marine algae of California: I.A. Abbott and G.J. Hollenberg. Stanford University Press, Stanford, California, 1976, xii +827 pp., 701 figures, 5 maps. Price 22.50 U.S.$, ISBN 0-8047-0867-3. Aquat Bot. 3:391–392.
- Druehl LD. 1978. The distribution of Macrocystis integrifolia in British Columbia as related to environmental parameters. Can J Bot. 56:69–79.
- Galtier N, Nabholz B, Glémin S, Hurst GDD. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18:4541
- Graham MH, Vásquez JA, Buschmann AH. 2007. Global ecology of the giant kelp Macrocystis: from ecotypes to ecosystems. Oceanogr Mar Biol. 45:39–88.
- Hoek CVD, Mann DG, Jahns HM. 1995. Algae: an introduction to phycology/C. van den Hoek, D.G. Mann, H.M. Jahns. Bull Mar Sci Miami. 59:242–243.
- Temple WD, Bomke AA, Radley RA, Holl FB. 1989. Effects of kelp (Macrocystis integrifolia and Ecklonia maxima) foliar applications on bean crop growth and nitrogen nutrition under varying soil moisture regimes. Plant Soil. 117:75–83.
