Abstract
Jinbian carp (Cyprinus carpio) is an endemic species in China. The complete mitochondrial genome of Jinbian carp is determined to be 16,581 bp in length and includes 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and a control region. Its structural organization and gene order are equivalent to other common carp strains. The phylogenetic analyses will contribute to further insights of the taxonomy and phylogeny in Cyprinidae family.
Common carp is old freshwater fish that have been cultured as aquaculture species all over the world for thousand years (FAO Citation2004). Jinbian carp (Cyprinus carpio) is an endemic species that mainly distributed in Guangxi Province, China. It has two golden stripes on both sides of its Dorsal fin, which can be used to distinguish from other carp varieties. Due to its high-protein, low-fat, easy culture, and fast growth, Jinbian carp was regularly cultured in rice paddy and ponds as an ecological friendly agriculture-aquaculture form in Southwest China. To better define the phylogenetic relationship between Jinbian carp and other closely related varieties, we cloned and analyzed its complete mitochondrial genome sequence for the first time.
In this study, the Jinbian carp was sampled from Rongshui (25°04′N, 109°14′E), Guangxi province, China. Fin tissues were preserved in 95% ethyl alcohol. The extracted DNA is stored in the laboratory of Aquatic Species Introduction and Breeding Center of Guangxi, China. Seventeen pairs of primers and DNA samples were used to amplify the complete mitogenome using PCR-based method (Mabuchi et al. Citation2006; Zhang et al. Citation2009).
The mitogenome sequence of Jinbian carp was 16,581 bp in length (GenBank accession No. MH202953) and consisted of 13 protein-coding genes (PCGs), 22 tRNAs, 2 rRNAs, and a control region (D-loop). The overall base composition was 31.92% A, 15.73% G, 24.82% T, and 27.53% C, with a slight A + T bias. In the 13 PCGs, ND6 gene especially stayed on the L-strand while other genes were on the H-strand. Most of PCGs started with ATG while CO1 genes used GTG as start codon. ND2 and ND3 genes were used TAA as the stop codon, ND4 and cytb genes were ended with incomplete stop codon of T. The structural organization and gene order of mtDNA sequence of Jinbian carp was similar to other common carp strains (Chang et al. Citation1994; Wang et al. Citation2013; Liu et al. Citation2016).
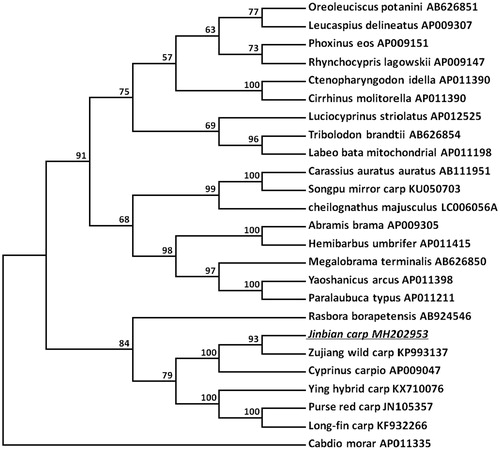
A phylogenetic tree was constructed to validate the phylogenetic position of Jinbian carp. MEGA package was used to construct a Neighbor-joining tree containing complete sequences of Jinbian carp and other 19 Cyprinidae varieties downloaded from GenBank (). Cabdio morar was used as an outgroup member. The tree showed that Jinbian carp was a group with other Cyprinus carpio in Cyprinidae family and closely related to Zujiang wild carp.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- FAO 2004. Yearbook of Fisheries Statistic Summary Tables. http://www.fao.org/fi/statistic/summtab/default.asp.
- Chang YC, Huang F, Lo TB. 1994. The complete nucleotide sequence and gene organization of carp (Cyprinus carpio) mitochondrial genome. J Mol Evol. 38:138–155.
- Mabuchi K, Miya M, Senou H, Suzuki T, Nishida M. 2006. Complete mitochondrial DNA sequence of the Lake Biwa wild strain of common carp (Cyprinus carpio L.): further evidence for an ancient origin. Aquaculture. 257:68–77.
- Wang B, Ji P, Wang J, Sun J, Wang C, Xu P, Sun X. 2013. The complete mitochondrial genome of the Oujiang color carp, Cyprinus carpio var. color (Cypriniformes, Cyprinidae). Mitochondrial DNA. 24:19–21.
- Zhang X, Yue B, Jiang W, Song Z. 2009. The complete mitochondrial genome of rock carp Procypris rabaudi, (Cypriniformes: Cyprinidae) and phylogenetic implications. Mol Biol Rep. 36:981.
- Liu XJ, Liang YJ, Lu CY, Chang YM, Li CT, Qin XF, Quan SY, Wang ZY, Zou GW, Hu GF. 2016. Mitochondrial genome of Ying hybrid carp (Russian scattered scale mirror carp ♀ × carp-goldfish nucleocytoplasmic hybrid ♂). Mitochondrial DNA B. 1:925–926.