Abstract
The entire mitochondrial genome of Cobitis nalbanti (Teleostei: Cypriniformes: Cobitidae) was analyzed using the primer walking method. The mitogenome was 16,631 bp in length, comprising 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. Its gene order was congruent with those of typical vertebrates. In the phylogenetic tree, C. nalbanti was clearly separated from C. lutheri, which supported the recent taxonomic revision.
Cobitis nalbanti (Teleostei: Cypriniformes: Cobitidae) in South Korea was previously reported as Cobitis lutheri, until Vasil’eva et al. (Citation2016) redescribed the latter and erected the former as a new species endemic to Korea due to its morphological and cytogenetic characteristics. In this study, the full-length mitochondrial genome of C. nalbanti was analyzed.
A specimen of C. nalbanti was collected at Jangpyeong-myeon, Cheongyang-gun in Korea in 2009. The voucher specimen (SCU-0009) was deposited in the aquatic animal collection of the Department of Life Sciences and Biotechnology, Soonchunhyang University (SCU; Asan, South Korea). The genomic DNA was extracted from its fin tissue (Asahida et al. Citation1996), and the mitogenome was amplified using two independent and overlapping PCR runs, followed by sequencing with a set of 27 primers. The complete sequence was deposited in the GenBank; the accession number MH349461.
Mitogenome sequences of the ‘northern clade’ of the subfamily Cobitinae (Šlechtová et al. Citation2008) were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/). They were aligned together with the C. nalbanti sequence and refined manually to correct obvious misalignments. The nucleotide matrix of 12 protein-coding genes, excluding nad6, was created with the first, second, and third bases of each codon. The final matrix comprised 3619 bp for each base. The alignment information is available upon request in FASTA format. Phylogenetic analysis was conducted using RAxML 7.0.4 (Stamatakis Citation2006) for maximum likelihood (ML) analysis.
The C. nalbanti mitogenome was a circular molecule of 16,631 bp in length, consisting of 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. The order of these genes was identical to those of corresponding genes that typical vertebrates have.
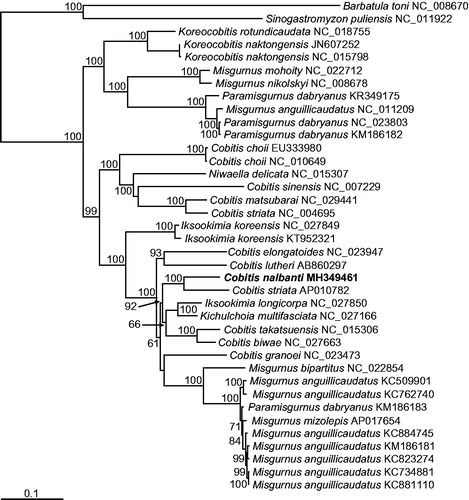
With this complete mitogenome sequence of C. nalbanti, a phylogenetic tree was reconstructed by the ML method, using the nucleotide sequence matrix from 12 concatenated protein-coding genes (). In the resulting tree, the cobitid species of the “northern clade” of the subfamily Cobitinae (Šlechtová et al. Citation2008) were divided into the two lineages. The first lineage included the genera Misgurnus and Paramisgurnus that did not experience mitochondrial introgression, and the two species of the genus Koreacobitis. The second lineage consisted of cobitid species belonging to the genera Misgurnus and Paramisgurnus that experienced mitochondrial introgression, and the genera Cobitis, Iksookimia, Kichulchoia, and Niwaellea, forming a strong monophyletic lineage overall. Among them, C. nalbanti, the newly erected species by Vasil’eva et al. (Citation2016) that was formally classified as C. lutheri was consistently separated from C. lutheri. Therefore, our results supported the taxonomic conclusion of Vasil’eva et al. (Citation2016). Interestingly, our phylogenetic tree supported the monophyletic status of neither Cobitis, Iksookimia, Kichulchoia, nor Niwaellea, which is clearly different from the multiple nuclear gene-based tree of Kwan et al. (Citation2018). In the future study, the mitochondrial introgression after hybridization among the species of those genera, as previously observed among some species in the genera Misgurnus and Paramisgurnus (Šlechtová et al. Citation2008), should be investigated to clarify their in-depth phylogenetic relationships and evolutionary history.
Figure 1. Maximum likelihood (ML) phylogeny based on the full-length mitochondrial genomes from the cobitid species belonging to the subfamily Cobitinae. The matrix included the three codon positions of the 12 protein-coding genes. A bootstrap value above 50% in the ML analysis is indicated at each node. Cobitis nalbanti analyzed in this study is shown in bold.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Asahida T, Kobayashi T, Saitoh K, Nakayama I. 1996. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentration of urea. Fish Sci. 62:727–730.
- Kwan YS, Kim D, Ko M, Lee W, Won Y. 2018. Multi-locus phylogenetic analyses support the monophyly and the Miocene diversification of Iksookimia (Teleostei: Cypriniformes: Cobitidae). Syst Biodiver. 16:81–88.
- Šlechtová V, Bohlen J, Perdices A. 2008. Molecular phylogeny of the freshwater fish family Cobitidae (Cypriniformes: Teleostei): delimitation of genera, mitochondrial introgression and evolution of sexual dimorphism. Mol Phylogenet Evol. 47:812–831.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690.
- Vasil’eva Ekaterinad, Kim Daemin, Vasil’ev Victorp, Ko Myeong-Hun, Won Yong-Jin, Vasil'eva E, Kim D, Vasil’ev V, Ko M-H, Wo Y-J. 2016. Cobitis nalbanti, a new species of spined loach from South Korea, and redescription of Cobitis lutheri (Teleostei: Cobitidae). Zootaxa. 4208:577–591.
