Abstract
A pika species Ochotona thibetana sacraria (Lagomorpha) (Thomas Citation1923), was reported as a subspecies of O. thibetana. To date, this subspecies have been elevated to species status. O. sacraria belongs to a new subgenus Alienauroa and is endemic to Mount Emei of Sichuan, China. In this study, the complete mitogenome of O. sacraria was determined. The mitogenome is a circular molecule of 16,573 bp in length, containing 13 protein-coding genes, 2 ribosome RNA genes, 1 light strand replication origin (OL), 22 transfer RNA genes and 1 non-coding region. We reconstructed a phylogenetic tree based on Bayesian inference for 12 small mammal species. The new mitogenome data would provide useful information for application in conservation genetics and further clarify phylogenetic evolution of this species.
Thomas (1923) named a specimen which was collected from Mount Emei in 1910 as O. thibetana sacraria. However, Allen (Citation1938) regarded it as synonym of Tibetan pika. Subsequently, Feng (Citation1973) restored it as a subspecies of O. thibetana. Now, this subspecies have been elevated to species status, and belonged to a new subgenus Alienauroa by Liu et al. (Citation2017). O. sacraria is endemic to Mount Emei, Sichuan, China. The species is associated with the forested habitats and distribute in low altitude (1,200–3,100 m) (Liu et al. Citation2017). To date, complete mitochondrial genome data of O. sacraria is not available in the GenBank. In this study, we sequenced the complete mitochondrial genome of O. sacraria (GenBank number: MH345726) and examined its phylogenetic position with other 11 small mammal species.
The tissue samples were obtained in Mount Emei, Sichuan province, China (Latitude: 29.54°N, Longitude: 103.33°E), and maintained in Sichuan Normal University, Chengdu. Total genomic DNA was extracted from liver tissue using the DNA extraction kit (Aidlab Biotech, Beijing, China). The mitochondrial genomes of O. erythrotis (NC_037186.1) is used to design primers for polymerase chain reaction (PCR) and used as template for gene annotation.
The total complete mitogenome sequence of O. sacraria is 16,473 bp, which is composed of 13 protein-coding genes (PCGs), 2 ribosome RNA genes, 1 light-strand replication origin (OL), 22 transfer RNA genes and 1 non-coding region. The total base composition of the O. sacraria mt genome is an A + T-rich pattern of the vertebrate mitochondrial genomes. ATG is the most common start codon, ATT and ATC is used for ND2, ND5 and ND3. Most tRNAs could be folded into the canonical cloverleaf secondary structure, except Trna-Ser(AGY). O. sacraria had two non-coding regions: a 35 bp L-strand replication origin (OL) and a 1,094 bp control region (D-loop). The major noncoding control region (D-Loop) had one tandem repeat element, including 46 copies of a 6 bp tandem repeat element (5′-GTACGT-3′).
The phylogenetic relationship for the mitochondrial genome sequences newly determined was examined with those of 4 Lepus, 4 Ochotona, and 3 outgroup species. The BI analysis was performed using BEAST v1.7 (Drummond et al. Citation2012), and the best-fit model (GTR + I + G) of nucleotide evolution was selected using the AIC test in JModelTest 2 (Darriba et al. Citation2012). Phylogenetic tree resulting from the Bayesian inference (BI) analyses showed that the species forms one basal position (posterior probability (pp) = 1.00) ().
Figure 1. Phylogenetic tree derived from 12 protein-coding gene sequences from 12 complete mitochondrial genomes using BI analysis. Numbers by the nodes indicate Bayesian posterior probabilities. Branch lengths are mean estimates.
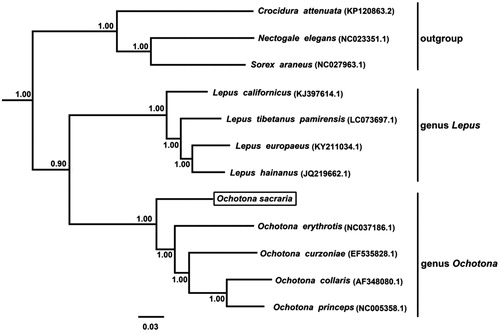
We first reported and analyzed the complete mitochondrial genome of O. sacraria. The data will contribute to solve the phylogenetic position of the genus Ochotona.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Allen GM. 1938. The mammals of China and Mongolia natural history of Central Asia. New York: The American Museum of Natural History.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. JModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973.
- Feng T. 1973. A new species of Ochotona (Ochotonidae, Mammalia) from Mount Jolmo-Lungma area. Acta Zoolog Sin. 19:69–75.
- Liu SY, Jin W, Liao R, Sun ZY, Zeng T, Fu JR, Chen LM. 2017. Phylogenetic study of Ochotona based on mitochondrial Cyt b and morphology with a description of one new subgenus and five new species. Acta Theriol Sin. 37:1–43.
- Thomas O. 1923. On mammals from the Li-kiang range, Yunan, being a further collection obtained by Mr. Georoge Forrest. Ann Mag Nat Hist. 11:655–663.
