Abstract
Caulerpa cupressoides (Vahl) C. Agardh is a widely distributed tropical green algae. The circular chloroplast genome was 130,895 bp in length, with a GC content of 34%. In total, 99 genes were identified and they were consisted of 63 coding genes, 30 tRNA genes, and 6 rRNA genes. This chloroplast genome did not show an obvious quadripartite structure. Phylogenetic analysis revealed that C. cupressoides, C. racemosa, and Tydemania expeditionis were close relatives, with high bootstrap values. The characterized complete chloroplast genome of C. cupressoides will provide essential date for further studies of Bryopsidales.
Caulerpa cupressoides (Vahl) C. Agardh is a siphonous green seaweed belonging to the genus Caulerpa and distributed in tropical seas (Nielsen and Price Citation2001). Recently, the antinociceptive and anti-inflammatory activities of the lectin isolated from this species have been proved (Benevides et al. Citation2001; Vanderlei et al. Citation2010; Da et al. Citation2014), indicating its potential medical value. In this study, we reported a complete cp genome of the C. cupressoides.
The fresh thallus of C. cupressoides was collected from Sanya (Hainan, China; 109°30′11.52″E, 18°12′36.72″N), and were used for the total genomic DNA extraction with the modified CTAB method (Doyle Citation1987). Voucher herbarium specimens were prepared and deposited in Marine Biological Museum of the Chinese Academy of Sciences (MBMCAS). The whole-genome sequencing was conducted with 150 bp pair-end reads on the Illumina Hiseq Platform (Illumina, San Diego, CA). In total, 2.3 million high quality base pairs of sequence data were obtained and the genome was assembled with NOVOPlasty (Dierckxsens et al. Citation2017). Protein-coding genes and non-coding RNAs were annotated with GeSeq (Tillich et al. Citation2017). The annotations chloroplast genome was submitted to GenBank database under Accession (No. MG797569). The phylogenetic relationship of C. cupressoides was conducted based on the complete chloroplast genome of 14 species in Trebouxiophyceae and Ulvophyceae. The complete cp genome sequences were aligned by MAFFT software (Katoh et al. Citation2002) and the phylogenetic tree was constructed using RAxML (Stamatakis Citation2014) by maximum-likelihood method.
The complete chloroplast genome of C. cupressoides was 130,895 bp in length. The chloroplast genome contained 99 genes, including 63 protein-coding genes, 30 transfer genes, and 6 ribosomal RNA genes. Like congeneric C. racemosa, this chloroplast genome did not show a quadripartite structure and lacked the large rRNA operon-encoding Inverted Repeat (IR). The number of tRNA and rRNA were same between C. cupressoides and C. racemose, while the number of genes was 7 less than C. racemose due to few ORFs genes. The overall base composition was 32.2% for A, 17.4% for C, 16.6% for G and 33.8% for T.
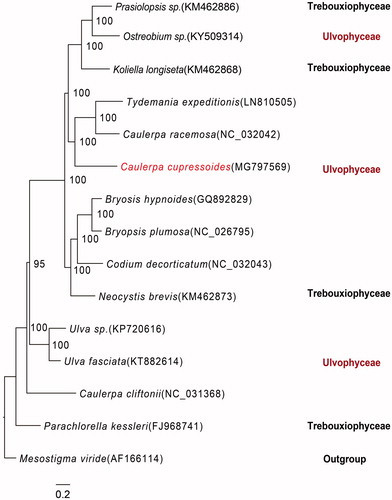
The phylogenetic tree analysis showed that C. cupressoides was clustered with T. expeditionis and C. racemose (). However, the species C. cliftonii of the same genus was not similar. Our results revealed that it was quite similar to previously published trees (Lam and Zechman Citation2006; Fučíková et al. Citation2014; Leliaert and Lopez-Bautista Citation2015; Melton III et al. Citation2015). To date, the classification of Trebouxiophyceae and Ulvophyceae was not clear enough compared to previous studies. The chloroplast genome traits have the potential to provide insights of deep-level relationships at the Bryopsidales level.
Acknowledgements
We thank Ruoyu Liu (Institute of Deep-sea Science and Engineering, Chinese Academy of Sciences) for his help during collection of specimens. We also thank Dr. Kun Wang for providing invaluable comments, which improved the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Benevides N, Holanda M, Melo F, Pereira M, Monteiro A, Freitas A. 2001. Purification and partial characterization of the lectin from the marine green alga Caulerpa cupressoides (Vahl) C. Agardh. Bot Mar. 44:17–22.
- Da CRR, Chaves HV, do Val DR, de Freitas AR, Lemos JC, Rodrigues JA, Pereira KM, de Araújo IW, Bezerra MM, Benevides NM. 2014. A lectin from the green seaweed Caulerpa cupressoides reduces mechanical hyper-nociception and inflammation in the rat temporomandibular joint during zymosan-induced arthritis. Int Immunopharmacol. 2014:34–43.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45:e18.
- Doyle JJ, Doyle JLJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Fučíková K, Leliaert F, Cooper ED, Škaloud P, D'Hondt S, De Clerck O, Gurgel CFD, Lewis LA, Lewis PO, Lopez-Bautista JM, et al. 2014. New phylogenetic hypotheses for the core Chlorophyta based on chloroplast sequence data. Front Ecol Evol. 2:63.
- Katoh K, Misawa K, Kuma Ki MT. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast fourier transform. Nucleic Acids Res. 30:3059–3066.
- Lam DW, Zechman FW. 2006. Phylogenetic analyses of the Bryopsidales (Ulvophyceae, Chlorophyta) based on Rubisco large subunit gene sequences. J Phycol. 42:669–678.
- Leliaert F, Lopez-Bautista JM. 2015. The chloroplast genomes of Bryopsis plumosa and Tydemania expeditiones (Bryopsidales, Chlorophyta): compact genomes and genes of bacterial origin. BMC Genomics. 16:204.
- Melton JT, III Leliaert F, Tronholm A, Lopez-Bautista JM. 2015. The complete chloroplast and mitochondrial genomes of the green macroalga Ulva sp. UNA00071828 (Ulvophyceae, Chlorophyta). PLoS One. 10:e0121020.
- Nielsen R, Price IR. 2001. Typification of Caulerpa cupressoides (Vahl) C. Agardh and C. taxifolia (Vahl) C. Agardh (Chlorophyta, Caulerpaceae). Taxon. 50:827–836.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes . Nucleic Acids Res. 45:W6–W11.
- Vanderlei ESO, Patoilo KKNR, Lima NA, Lima APS, Rodrigues JAG, Silva LMCM, Lima MEP, Lima V, Benevides NMB. 2010. Antinociceptive and anti-inflammatory activities of lectin from the marine green alga Caulerpa cupressoides. Int Immunopharmacol. 10:1113–1118.