Abstract
Macrophthalmus japonicus is located in the Macrophthalmidae family, but genetic research about the whole mitochondrial genome in Macrophthalmidae family doesn’t exist. Here, we report the first complete mitochondrial genome from M. japonicus, which is composed of 16,170 base pair (bp) containing 13 protein coding genes, 22 transfer RNAs, 2 ribosomal RNAs, and A + T rich region. The overall nucleotide composition of M. japonicus was G + C: 33.6%, A + T: 66.4% with a strong AT bias. The phylogenetic relationship using whole mitochondrial genome showed that M. japonicus was closest with Eriocheir japonica although their respective families are different.
In morphological classification, the Macrophthalmus japonicus was classified in the Decapoda order and the Macrophthalmidae family. The M. japonicus lives in mudflats and are also called as mud crabs. It is known that the burrows in mudflats can supply oxygen and play a role in organic carbon cycle (Otani et al. Citation2010). There is a research that lectin from M. japonicus can be applied for the detection of malignant tumor or cancer cell in humans (Kim et al. Citation2006), because of this the M. japonicus may have a value as biological resource. However, there is no research related to whole mitochondrial genome (mitogenome) in the Macrophthalmidae family. Therefore, we studied about the mitogenome of M. japonicus and investigated if there was phylogenetic relationship with other crabs in the Decapoda order. Samples of M. japonicus were taken from Gangseo-gu, Busan, Republic of Korea (GIS: 35°04′47′′N 128°52′36′′E). A specimen of this crab can be found at the National Institute of Biological Resources, Republic of Korea, under the certificate number NIBRGR0000114816. The muscle tissue was grinded and the mitochondria were harvested by differential centrifugation (Graham Citation2001). QIAamp Tissue kit (Qiagen, CA, USA) was used to extract the mitogenomic DNA. After amplification of the mitochondrial DNA by PCR, NGS library preparation and sequencing by Ion Torrent PGM platform were done based on the manufacturer’s instructions (Life Technologies/Thermo Fisher Scientific, Waltham, MA, USA). The mitogenome assembly and gene annotation were performed using CLC genomics workbench 8.5 (CLC Bio, Denmark). Annotation results in protein coding gene (PCGs), ribosomal RNA (rRNAs), and transfer RNA (tRNAs) were confirmed based on the NCBI Basic Local Alignment Search Tool (BLAST) and tRNA-scan 1.21 (Altschul et al. Citation1990; Lowe and Eddy Citation1997). To figure out phylogenetic relationship of M. japonicas with other crabs in the order Decapoda, a phylogenetic tree was illustrated using the neighbor-joining method with 1000 replicate bootstrap by MEGA 6.0 (Tamura et al. Citation2013). In the phylogenetic tree, the mitogenome from other crabs in Decapoda order was used.
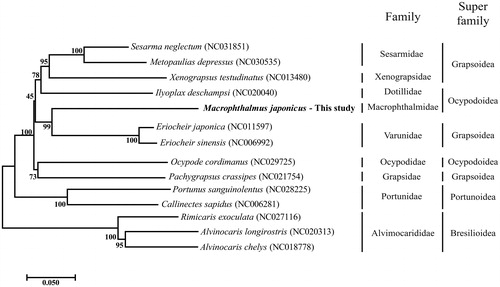
The complete mitogenome sequence of M. japonicus consists of circular DNA with 16,170 bp containing 13 protein coding gene (PCGs), 22 transfer RNA (tRNAs), 2 ribosomal RNA (rRNAs), and A + T rich region. The sequence was submitted to GenBank (accession number: NC030048). In 13 PCGs, three start codons such as ATG, ATT and ATA were used. Stop codon TAA and TAG were used in most of PCGs except nd6, cytb, cox1, and cox3. In these four PCGs, incomplete stop codon T (aa) or TA (a) were used for translation termination (Yamauchi et al. Citation2003; Hou et al. Citation2007; Zhang et al. Citation2016). The nucleotide composition was asymmetric (A: 33.6%, T: 32.7%, G: 11.0%, C: 22.7%) with a strong AT bias. In the phylogenetic tree, M. japonicus was closest to Eriocheir japonica (NC011597) which is located in a different family (Varunidae) (). This result emphasizes that mitogenome can be a way for identification of the crab in Decapoda order.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Graham JM. 2001. Current Protocols in Cell Biology: isolation of Mitochondria from Tissues and Cells by Differential Centrifugation. Liverpool (LIV). John Wiley & Sons, Inc. 3:1–15.
- Hou W, Chen Y, Wu X, Hu J, Peng Z, Yang J, Tang Z, Zhou C-Q, Li Y-m, Yang S. 2007. A complete mitochondrial genome sequence of Asian black bear sichuan subspecies (Ursus thibetanus mupinensis). Int J Biol Sci. 3:85–90.
- Kim H-N, Chung W-H, Bae C-H, Hwang K-W, Kim H-H. 2006. Isolation and carbohydrate binding specificity of a lectin from the hemolymph of coastal crab Macrophthalmus japonicus. Yakhak Hoeji. 50:166–171.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Otani S, Kozuki Y, Yamanaka R, Sasaoka H, Ishiyama T, Okitsu Y, Sakai H, Fujiki Y. 2010. The role of crabs (Macrophthalmus japonicus) burrows on organic carbon cycle in estuarine tidal flat, Japan. Estuar Coast Shelf Sci. 86:434–440.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30:2725–2729.
- Yamauchi MM, Miya MU, Nishida M. 2003. Complete mitochondrial DNA sequence of the swimming crab, Portunus trituberculatus (Crustacea: Decapoda: Brachyura). Gene. 311:129–135.
- Zhang Q-X, Guan D-L, Niu Y, Sang L-Q, Zhang X-X, Xu S-Q. 2016. Characterization of the complete mitochondrial genome of the Asian plant hopper Ricania speculum (Hemiptera: Fulgoroidea: Ricannidae). Conservation Genet Resour. 8:463–466.