Abstract
Macrothelypteris torresiana is a reputed medicinal fern. Its complete chloroplast genome was determined by Illumina paired-end sequencing. The genome is 151,150 bp in length with 43.1% overall Guanine+Cytosine (GC) content, which is divided into four distinct parts such as a small single copy (SSC, 21,772 bp), a large single copy (LSC, 82,422 bp), and two inverted repeats (IRs, 23,478 bp each). It contains 132 genes, including 86 protein-coding genes, eight ribosomal RNA genes, 35 tRNA genes, and three pseudogenes. Maximum likelihood (ML) tree revealed that M. torresiana was closely grouped with Christella appendiculata with 100% bootstrap value.
Macrothelypteris torresiana (Gaud.) Ching is a robust fern belonging to Thelypteridaceae. Its distinguished features include the glaucous-white, almost glabrous young stripes prior to the fronds unfurling (de Lange and Crowcroft Citation1997). The species prefers wet places in mountain valleys at an altitude of 1000 m with distribution in tropical and subtropical regions including China, America, Australia, and Pacific islands (Lin et al. Citation2013). As a reputed folk medicinal fern, its leaves and roots are widely used for fever, pain, and granulation in Southeast Asia and for kidney problems in China (Huang et al. Citation2010; Chen et al. Citation2012). The species is suffering different classified process including assigning to Macrothelypteris, Phegopteris, and allied genera (Smith and Cranfill Citation2002; He and Zhang Citation2012). Hence, the acquirement of whole chloroplast (cp) genome of M. torresiana will be helpful to deeply investigate phylogenetic relationships of Thelypteridaceous genera.
We obtained mature and healthy leaves of M. torresiana from South China Botanical Garden, Chinese Academy of Sciences (23°11′3.56″N, 113°21′43.28″E), which was used to DNA extraction through Tiangen Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China). The specimen is stored in the Herbarium of Sun Yat-sen University (SYS; voucher: SS Liu 201618). After DNA was broken into 300 bp fragment using Covaris M220 (Covaris Inc., Woburn, MA), an Illumina paired-end (PE) genomic library was constructed and genome sequencing was performed on Hiseq 2500 platform (Illumina Inc., San Diego, CA). In total, 2.04 G raw reads were quality-trimmed by Trimmomatic v0.32 (Bolger et al. Citation2014) and 1.74 G clean data were used to assemble the chloroplast genome by Velvet v1.2.07 (Zerbino and Birney Citation2008). We used Dual Organellar GenoMe Annotator (DOGMA; Wyman et al. Citation2004) to annotate genes and tRNAscan-SE programs (Lowe and Eddy Citation1997) to confirm tRNAs. In order to survey phylogenetic classification, we selected the complete chloroplast genome sequence of 11 ferns including M. torresiana with Angiopteris evecta as an outgroup and created a multiple sequence alignment by the MAFFT v7.311 (Katoh and Standley Citation2013). A maximum likelihood (ML) tree was inferred using RAxML v.8.0 (Stamatakis Citation2014) with 1000 bootstrap replicates.
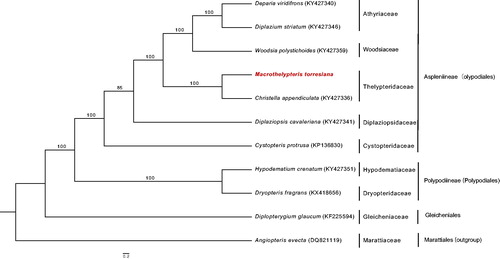
The complete chloroplast genome of M. torresiana is a double-stranded circular DNA of 151,150 bp in length with 43.1% overall GC content (GenBank Accession Number: MH500230), which was divided into four distinct parts such as a small single copy (SSC) of 21,772 bp, a large single copy (LSC) of 82,422 bp, and two inverted repeats (IRa and IRb) of 23,478 bp. The GC content of the LSC, SSC, and IR are 42.6, 40.2, and 45.3%, respectively. The chloroplast genome contains 132 genes, including 86 protein-coding genes, eight ribosomal RNA genes, 35 tRNA genes, and three pseudogenes, and 14 genes duplicated in the IR region. Eighteen genes have one or two introns involving ndhB, rps16, atpF, rpoC1, petB, petD, ndhA, rpl16, rpl2, trnG-UCC, trnV-UAC, trnA-UGC, trnI-GAU, trnL-UAA, trnT-UGU, ycf3, clpP, and rps12. The ML tree showed that M. torresiana was closely grouped with Christella appendiculata with 100% bootstrap value (). The complete chloroplast genome of M. torresiana will provide powerful data for facilitating phylogenomics of ferns.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. http://www.usadellab.org/cms/?page=trimmomatic
- Chen JL, Lei YF, Wu GH, Zhang YH, Fu W, Xiong CM, Ruan JL. 2012. Renoprotective potential of Macrothelypteris torresiana via ameliorating oxidative stress and proinflammatory cytokines. J Ethnopharmacol. 139:207–213.
- de Lange PJ, Crowcroft GM. 1997. Macrothelypteris torresiana (Thelypteridaceae) at North Cape, North Island, New Zealand – a new southern limit for a tropical fern. New Zeal J Bot. 35:555–558.
- He LJ, Zhang XC. 2012. Exploring generic delimitation within the fern family Thelypteridaceae. Mol Phylogenet Evol. 65:757–764.
- Huang XH, Xiong PC, Xiong CM, Cai YL, Wei AH, Wang JP, Liang XF, Ruan JL. 2010. In vitro and in vivo antitumor activity of Macrothelypteris torresiana and its acute/subacute oral toxicity. Phytomedicine. 17:930–934.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. https://mafft.cbrc.jp/alignment/software/index.html
- Lin YX, Li ZY, Iwatsuki K, Smith AR. 2013. Thelypteridaceae. In: Wu ZY, Raven PH, Hong DY, eds. Flora of China, Vol. 2–3 (Pteridophytes). Beijing: Science Press; p. 319–396.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964.
- Smith AR, Cranfill RB. 2002. Intrafamilial relationships of the Thelypteroid ferns (Thelypteridaceae). Am Fern J. 92:131–149.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. https://sco.h-its.org/exelixis/web/software/raxml/
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255. http://dogma.ccbb.utexas.edu/
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829. https://www.ebi.ac.uk/~zerbino/velvet/