Abstract
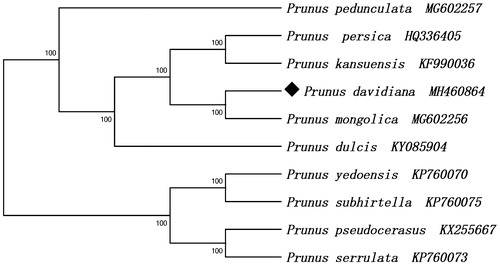
Prunus davidiana is a Chinese wild peach species belonging to the family Rosaceae. In this study, we first assembled the complete chloroplast (cp) genome of P. davidiana by Illumina paired-end reads data, then carried out phylogenetic analysis. The P. davidiana cp genome was 158,055 bp, containing a large single copy region (LSC) of 86,248 bp and a small single copy region (SSC) of 19,047 bp separated by a pair of inverted repeats (IRs) of 26,380 bp. The cp genome contained 131 genes, including 86 protein-coding genes, 37 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. The result of the phylogenetic analysis showed that P. davidiana is closely related to P. mongolica.
Prunus davidiana is a wild species, belonging to the subfamily Prunoideae of the family Rosaceae. It is native to China and is naturally distributed in most of the provinces of China and exists in various habitats (Leng and Qi Citation2003). Prunus davidiana possesses the advantages of highly drought-resistant and survival rates (Dang et al. Citation2017), therefore it is very suitable for extensive cultivation and also can reduce the rate of soil erosion in ecological restoration region (Liu et al. Citation2018). Prunus davidiana is related to the cultivated peach, and is a source of abundant resistance to insect pests and diseases (Smykov et al. Citation1982). Accordingly, this plant has the potential to be used as an advantage traits gene donor source which can be of benefit for future study in peach breeding programs. In this paper, we characterized the complete chloroplast genome sequence of P. davidiana, to contribute in further population genetic studies of the genus Prunus of the family Rosaceae.
Fresh leaves of P. davidiana were collected from Yulin, Shaanxi, China (37°51'N, 110°23'E). A voucher specimen (AF-06-22) was stored in the Institute of College of Horticulture, Northwest A&F University, Yangling, China. Genomic DNA was isolated with the CTAB method (Doyle and Doyle Citation1987). The obtained chloroplast DNA was sequenced using Illumina HiSeq 2500 Sequencing system (Illumina, CA, USA) and sequences were used for constructing library. In total, 14,666,376 of 150 bp raw paired-end reads were obtained. The program MITObim v1.8 (Hahn et al. Citation2013) was employed for the assembly of chloroplast genome, with that of Prunus persica (GenBank accession HQ336405) as the reference. Annotation was performed with Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004), and was manually inspected to predict PCGs, transfer RNA (tRNA) genes, and ribosomal RNA (rRNA) genes. The complete cpDNA sequence of P. davidiana has been submitted to GenBank with the accession number MH460864.
The complete chloroplast genome of P. davidiana was 158,055 bp in length, consisting of a pair of IR regions (26,380 bp), a small single copy (SSC) region (19,047 bp) and a large single copy (LSC) region (86,248 bp). The chloroplast genome of P. davidiana contained 131 genes including 86 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Most of these genes occurred as a single copy, however, eight protein-coding genes (ndhB, rpl2, rpl23, rps7, rps12, rps19, ycf1, ycf2), seven tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC) and four rRNA genes (4.5S, 5S, 16S, 23S) in the IR regions are totally duplicated. There were fifteen genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) containing one intron and three genes (ycf3, clpP, rps12) containing two introns. The overall GC content of P. davidiana chloroplast genome is 36.8% and the corresponding values in LSC, SSC, and IR regions are 34.6%, 30.3%, and 42.6%, respectively. The maximum likelihood (ML) tree showed that P. davidiana is closely related to P. mongolica ().
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Leng P, Qi JX. 2003. Effect of anthocyanin on David peach (Prunus davidiana Franch) under low temperature stress. Sci Hort. 97:27–39.
- Dang X, Liu G, Zhao L, Zhao G. 2017. The response of carbon storage to the age of three forest plantations in the Loess hilly regions of China. Catena. 159:106–114.
- Liu D, Huang Y, Yan H, Jiang Y, Zhao T, An S. 2018. Dynamics of soil nitrogen fractions and their relationship with soil microbial communities in two forest species of northern China. PLoS One. 13:e0196567.
- Smykov VK, Ovcharenko GV, Perfilyeva ZN, Shoeferistov EP. 1982. Estimation of the peach hybrid resources by its mildew resistance against the infection background. Byull Gos Nikitsk Bot Sada. 88:74–80.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41:e129–e129.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20:3252–3255.