Abstract
The complete mitochondrial genome sequence of Lagocephalus sceleratus was determined. The complete mitochondrial genome was 16,444 bp in length and contained 13 protein-coding genes, 22 transfer RNA genes, two ribosomal RNA genes, and two non-coding region (the control region and the origin of light strand replication). The overall base composition was A 27.59%, C 31.43%, G 17.12%, T 23.85%. All protein-coding genes started with an ATG initiation codon, except COI used GTG. With the exception of ND6 and eight tRNA genes, all other genes were encoded on the heavy strand. Additionally, the phylogenetic relationship of 37 Tetraodontidae species based on the complete genome was analyzed, and the result showed that L. sceleratus was clustered with other Lagocephalus species. These results would be useful for the investigation of phylogenetic relationship, taxonomic classification and phylogeography of the Tetraodontidae.
Lagocephalus sceleratus (Tetraodontiformes, Tetraodontidae) is widely distributed in the tropical Indian and Pacific Oceans (Okan Akyo Citation2005; Türker-Çakır et al. Citation2009). It is a Lessepsian invasive and poisonous fish species introduced into the Mediterranean Sea through the Suez Canal (Bentur et al. Citation2008; Bakopoulos et al. Citation2017). Furthermore, the tetrodotoxin (TTX) of L. sceleratus can be used in the pharmaceutical industry (Nader M. 2012). In order to better understand the classification and biological value of this species, we determined the complete mitochondrial genome (mitogenome) of L. sceleratus (GenBank accession number: MH550879) and constructed a phylogenetic tree of Tetraodontida. This study is expected to contributing to the classification, systematic evolution of L. sceleratus and further phylogenetic relationship of Tetraodontidae and Tetraodontiformes.
L. sceleratus was collected from the South China Sea (22°16'32”N 114°09'56”E) and stored in a refrigerator of −80 °C with accession number 20171015XG04. The specimen was identified based on the morphologic features and COI gene. Total genomic DNA was extracted from muscle tissue using the phenol-chloroform method (Barnett & Larson Citation2012) and then used for PCR amplification and sequencing. The phylogenetic analysis of 37 Tetraodontidae was conducted based on the neighbor-joining (NJ) method using MEGA5 with 10,000 bootstrap replication (Tamura et al. Citation2011).
The complete mitogenome sequence of L. sceleratus was 16,444 bp in length, consisting of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, two ribosomal RNA genes (12S rRNA and 16S rRNA) and two non-coding region (the control region and the origin of light strand replication). Except ND6 and eight tRNAs (Gln, Ala, Asn. Cys, Tyr, Ser, Glu, Pro), other genes were encoded on the heavy strand. The overall base composition was A 27.59%, C 31.43%, G 17.12%, T 23.85%. The gene arrangement was consistent with other Tetraodontidae mitochondrial genomes (Yue et al. Citation2009; Jiang et al. Citation2014; Liu et al. Citation2014; Gong et al. Citation2016; Li et al. Citation2016). Twelve protein-coding genes started with an ATG initiation codon, COI used GTG as an initiation codon. For the termination codon, eight protein-coding genes (ND1, ND2, ATP8, ATP6, COIII, ND4L, ND5, Cytb) ended with TAA, three protein-coding genes (COI, COII, ND4) with a single T, ND3 with TAG and ND6 with AGG. The 13 protein-coding genes were 11,397 bp in length, accounting for 69.31% of the complete mitogenome, which encodes 3,799 amino acids in total. The 12S rRNA was located between tRNAPhe and tRNAVal and the 16S rRNA was located between tRNAVal and tRNALeu. They were 948 bp and 1,673 bp in length, respectively. The control region, located between tRNApro and tRNAPhe, is 955 bp. The origin of light strand replication was located between tRNAAsn and tRNACys with 51 bp in length.
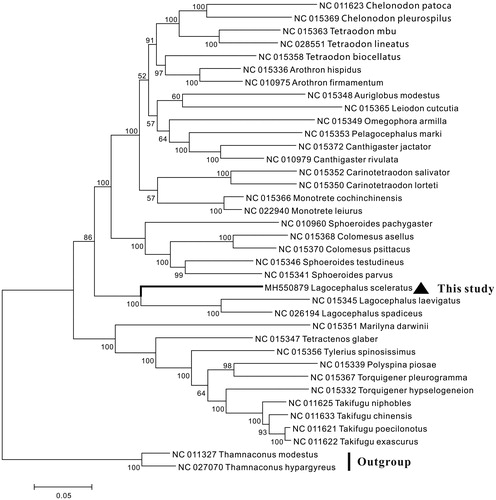
The phylogenetic relationship of L. sceleratus and other 36 Tetraodontidae species was constructed using the neighbor-joining (NJ) method based the complete mitogenome. The result showed that L. sceleratus first clustered with other two Lagocephalus species, Lagocephalus laevigatus and Lagocephalus spadiceus, suggesting L. sceleratus had a closer relationship with Lagocephalus species ().
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
Reference
- Bakopoulos V., Karoubali I. & Diakou A. (2017) Parasites of the Lessepsian invasive fish Lagocephalus sceleratus (Gmelin 1789) in the eastern Mediterranean Sea. Journal of Natural History 51, 421–34.
- Barnett R. & Larson G. (2012) A phenol-chloroform protocol for extracting DNA from ancient samples. Methods Mol Biol 840, 13–9.
- Bentur Y., Ashkar J., Lurie Y., Levy Y., Azzam Z.S., Litmanovich M., Golik M., Gurevych B., Golani D. & Eisenman A. (2008) Lessepsian migration and tetrodotoxin poisoning due to Lagocephalus sceleratus in the eastern Mediterranean. Toxicon 52, 964-8.
- Gong X., Zhang Q. & Bao B.-L. (2016) Complete mitochondrial DNA sequence of globe fish Tetraodon lineatus (Linnaeus, 1758) and the phylogenetic analysis of tetraodontidae. Mitochondrial DNA Part B 1, 781–2.
- Jiang H.-B., Bao J. & Han Y. (2014) Mitochondrial DNA sequence of the hybrid ofTakifugu flavidus(♀) × Takifugu rubripes(♂). Mitochondrial DNA, 1–2.
- Li X., Wu Z., Zhang G., Zhao C., Zhang H., Qian X. & Yin S. (2016) The complete mitochondrial genome of the hybrid of Takifugu fasciatus (♀) × T. flavidus (♂). Mitochondrial DNA Part B 1, 110–1.
- Liu Y., Zhou Q., Liu H., Li C. & Tong A. (2014) The complete mitochondrial genome sequence ofTakifugu flavidus(Tetraodontiformes: Tetrodontidae). Mitochondrial DNA 27, 613–4.
- Nader M. I.S., Boustany L. (2012) The Puffer Fish Lagocephalus sceleratus (Gmelin, 1789) in the Eastern Mediterranean. p. 38pp, FAO EastMed.
- Okan Akyo V.Ü., T. Tevfik Ceyhan and M. Bilecenoglu (2005) First confirmed record of Lagocephalus sceleratus (Gmelin, 1789) in the Mediterranean Sea. Journal of Fish Biology 66, 1183–6.
- Türker-Çakır D., Yarmaz A. & Balaban C. (2009) A new record ofLagocephalus sceleratus(Gmelin 1789) confirming a further range extension into the northern Aegean Sea. Journal of Applied Ichthyology 25, 606–7.
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M. & Kumar S. (2011) MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28, 2731–9.
- Yue G.H., Lo L.C., Zhu Z.Y., Lin G. & Feng F. (2009) The complete nucleotide sequence of the mitochondrial genome ofTetraodon nigroviridis. DNA Sequence 17, 115–21.