Abstract
Boehmeria is an important genus; however, no plastid genome has been reported to date. Here we report the complete chloroplast genomes for two Boehmeria species. The chloroplast genomes of Boehmeria umbrosa and Boehmeria spicata were found to be 170920 bp and 170958 in length, respectively, and the GC contents were 35.5 and 35.3%, respectively. The sequences of each species contained 112 unique genes, including 30 tRNA, 4 rRNA, and 78 protein-coding genes. This is the first report of cp genomes for Boehmeria, and will be useful for identifying molecular markers with which to address taxonomic problems in the genus.
Boehmeria Jacquin (Urticaceae) comprises approximately 47 species and is widely distributed in tropical and temperate regions (Wilmot-Dear and Friis Citation1996, Citation2013). It is an economically important genus which provides high-quality fibre (Chen et al. Citation2003). However, relationships within the genus still remain poorly resolved.
Information from chloroplast genomes has been extensively applied in understanding plant relationship (Ma et al. Citation2014; Du et al. Citation2017). To date, however, no complete plastid genome has been reported for any member of the Urticaceae.
In this study, we report and characterize the complete chloroplast genomes of Boehmeria umbrosa (Hand.-Mazz.) W. T. Wang and Boehmeria spicata (Thunberg) Thunberg, which are endemic to China and East Asia, respectively. Young, fresh, and healthy leaves were collected from B. umbrosa on Gaoligong Mountain (Yunnan, China; N 27°46′25.7″ E 98°35′38.35″), and from B. spicata on Tianmu Mountain (Zhejiang, China; N 30°20′14.8″, E 119°26′43.9″). Both voucher specimens were deposited in herbarium KUN (collection numbers are GLGE14989 and liuj10748, respectively). Genomic DNA was extracted following CTAB method (Doyle Citation1987), then sequenced using the Illumina Hiseq 4000. Sequences were assembled by multiple steps, including de novo assembling which was constructed in SPAdes version 3.9.1 (Bankevich et al. Citation2012), using k-mer lengths of 85–115 bp; then we used reference guided assembling conducted with Bandage version 0.8.1 (Wick et al. Citation2015) and Geneious version 9.1.4 (Kearse et al. Citation2012); Morus notabilis (NC_027110) was used as reference for assembling and annotation; finally, inverted repeat boundaries were determined by blast, and verified by reads mapping in Geneious version 9.1.4 (Kearse et al. Citation2012).
The complete chloroplast genome sequence of B. umbrosa (GenBank accession number MF990291) was 170920 bp in length, the GC content was 35.5%. LSC and SSC contained 68844 bp and 18462 bp, respectively, while IR was 41807 bp in length. The genome contained 112 functional genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
The complete chloroplast genome sequence of B. spicata (GenBank accession number MF990290) was 170958 bp in length, the GC content was 35.5%. LSC and SSC contained 70994 and 18478 bp, respectively, while IR was 40743 bp in length. The genome contained 112 functional genes, including 78 protein-coding genes, 30 tRNA genes, and 4 rRNA genes.
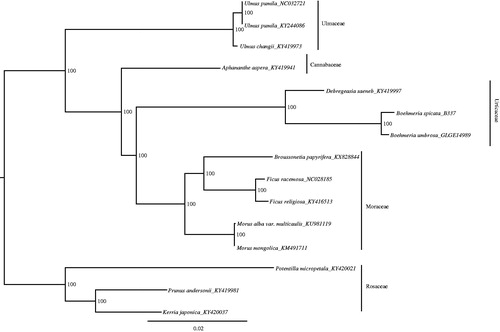
To identify the phylogenetic positions of B. spicata and B. umbrosa, a maximum likelihood phylogenetic tree was generated using RAxML–HPC BlackBox (Stamatakis Citation2014) through Cipres Science Gateway (Miler et al. Citation2010), based on concatenated complete chloroplast genomes from the two Boehmeria species, one Debregeasia species (GenBank-KY419997, which is only partial plastid genome) and other 12 species from Moraceae, Ulmaceae, Cannabaceae, and Rosaceae. Consistent with our previous results (Wu et al. Citation2013), present results showed that two species grouped into one well-supported clade and formed a sister to Debregeasia (). These newly characterized chloroplast genomes of Boehmeria can be used to develop markers for further study on the phylogeny and evolution of the genus Boehmeria, and also to clarify species boundaries, which is important for conservation.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Chen CJ, Lin Q, Friis I, Wilmot-Dear CM, Monro AK. 2003. Urticaceae. In: Wu ZY, Raven PH, editors. Flora of China. Beijing: Science Press & Beijing & Missouri Botanical Garden Press; p. 76–189.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Du YP, Bi Y, Yang FP, Zhang MF, Chen XQ, Xue J, Zhang XH. 2017. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci Rep. 7:5751.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Ma PF, Zhang YX, Zeng CX, Guo ZH, Li DZ. 2014. Chloroplast phylogenomic analyses resolve deep-level relationships of an intractable bamboo tribe Arundinarieae (Poaceae). Syst Biol. 63:933–950.
- Miler MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE); Nov 14, New Orleans (LA). p. 1–8.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31:3350–3352.
- Wilmot-Dear CM, Friis I. 1996. The New World species of Boehmeria and Pouzolzia (Urticaceae, tribus Boehmerieae). A taxonomic revision. Opera Bot. 129:1–103.
- Wilmot-Dear CM, Friis I. 2013. The old World species of Boehmeria (Urticaceae, tribus Boehmerieae). A taxonomic revision. Blumea Biodivers Ecol Biogeogr Plants. 58:85–216.
- Wu ZY, Monro AK, Milne RI, Wang H, Yi TS, Liu J, Li DZ. 2013. Molecular phylogeny of the nettle family (Urticaceae) inferred from multiple loci of three genomes and extensive generic sampling. Mol Phylogenet Evol. 69:814–827.