Abstract
The complete mitochondrial genome of Solegnathus hardwickii was determined to be 16,519 bp long circular molecule with a typical gene arrangement of vertebrate mitochondrial. The complete mitochondrial genomes were obtained by conventional and long PCR. Tree constructed using maximum likelihood based on protein-coding genes and ribosomal RNAs showed close relationship of S. hardwickii with Hippocampus spp.
Pallid seahorse Solegnathus hardwickii (Gray 1830) is a species of Syngnathidae family that distributed in habitats with hard substrates of Japan, Australia (eastern and northwestern) and South China Sea (Pollom Citation2017). The species of Solegnathus are highly worthwhile in the Asian medicine especially in Chinese traditional medicine (Courtney et al. Citation2007). Despite their valuable economic importance, there is a lack of information on the biology and ecology of Solegnathus species (Courtney et al. Citation2007). Here, we reported the complete mitochondrial genome of S. hardwickii (GenBank accession no. MH539788) to compare its phylogenetic platform with other members of Syngnathidae.
An adult of S. hardwickii was collected from South China Sea (Hainan province, China; 19° 02’ N 110° 44’ E) and stored in Hainan Tropical Ocean University Museum of Zoology (NO.0001-Sh). The genomic DNA was extracted from dorsal-lateral muscles (30 mg) using Rapid Animal Genomic DNA Isolation Kit (Sangon Biotech Co., Ltd., Shanghai, CN; NO. B518221). A genomic library was established followed by next-generation sequencing. Quality check for sequencing data was done by FastQC (Andrews Citation2010) and the fragments sequences were assembled and mapped using SPAdes version 3.9.0 (Petersburg, Russia) (Bankevich et al. Citation2012).
The complete sequence of S. hardwickii was 16,519 bp in size with a base composition of 29.75% A, 15.16% G, 26.73% T, and 28.36% C, including 13 protein coding genes (PCGs), 2 ribosomal RNAs (srRNA and lrRNA), 22 transfer RNAs (tRNAs), and and a control region (CR) of 903 bp. The genes of the S. hardwickii mitogenome are in the same order and orientation as in Doryichthys boaja mitogenome (Asem et al. Citation2018).
There was a strong A + T bias (56.47%). The longest gap and overlapping were determined between tRNA-Asn/tRNA-Cys (38 bp) and ATP8/ATP6 (10 bp), respectively. All PCGs began with common Met start codon, while COX1 was encoded with Val. Stop codons included TAA (ND1, ND2, COX1, ATP8, ATP6, ND4L and ND5), TAG (ND6) and incomplete codon T (COX2, COX3, ND3, ND4 and Cytb). The 12S ribosomal RNA and 16S ribosomal RNA were encoded from 71 to 1010 (940 bp) and 1084–2751 (1668 bp), respectively, with 16S having a rather higher A + T content (56.71 vs. 53.83%). These were located between the tRNA-Phe and tRNA-Leu, and were separated by the tRNA-Val. Seven tRNAs (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, and tRNA-Glu) and just ND6 protein-coding gene were encoded on the light strand and others were encoded on the heavy strand.
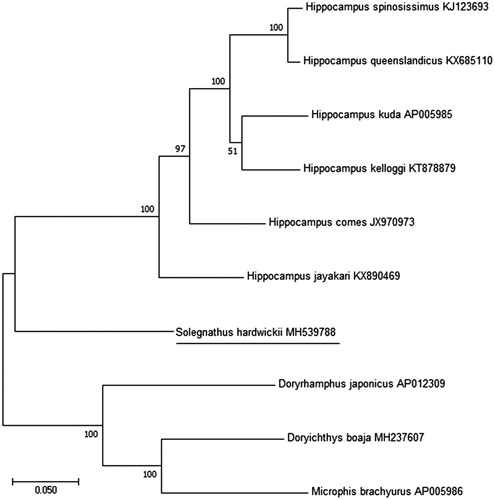
The phylogenetic relationship of S. hardwickii among Syngnathidae family was determined from a concatenated dataset including the 2 rRNAs and 13 PCGs using the software MEGA 7.0.26 version (Pennsylvania, USA). (Kumar et al. Citation2016) with 1000 bootstrap replicates and GTR model (). According to the result of phylogenetic tree, S. hardwickii was placed as a clade sister to Hippocampus spp. It needs to sequence the mitogenomes of other members of Syngnathidae to determine the phylogenetic status among them.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the manuscript.
Additional information
Funding
References
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- Asem A, Wang P, Li W. 2018. The complete mitochondrial genome of the Asian river pipefish Doryichthys boaja (Actinopterygii; Syngnathiformes; Syngnathidae) obtained using next-generation sequencing. Mitochondrial DNA. 3:776–777.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Courtney AJ, Haddy JA, Campbell MJ, Roy DP, Tonks ML, Gaddes SW, Chilcott KE, O’Neill MF, Brown IW, McLennan M, et al. 2007. An overview of the elasmobranch By-catch of the Queensland east coast trawl fishery (Australia). Australia: Department of Primary Industries and Fisheries Brisbane.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Pollom R. 2017. Solegnathus hardwickii. The IUCN Red List of threatened species. 2017:e.T20315A67623640.