Abstract
The parasites from buffaloes in Guizhou province, southwest China, were suspected to be sarcocysts through morphologic observation. 18S rRNA and the subunit I of cytochrome oxidase (cox1) sequences were amplified, then implemented by phylogenetic analysis. Phylogenetic analyses of 18S rRNA and cox1 genes both indicated the low degree of genetic diversity among the isolated sequences and other sequences of Sarcocystis fusiformis. The result shows that the sarcocysts from these buffaloes in Guizhou province S. fusiformis. This study is the first molecular characterization of S. fusiformis from Chinese buffaloes that used two genetic markers (18S rRNA and cox1 sequences) in China.
Introduction
Sarcocystis spp. have strict host demand that their development needs to be completed in two hosts (Dubey et al., Citation1989). The parasites reproduce asexually in their intermediate hosts, such as herbivores, omnivorous animals, and birds (Dahlgren & Gjerde, Citation2008; Lindsay et al., Citation2017). Then the protozoan would finish the sexual part of their life cycle in the definitive hosts which are carnivorous animals like dogs, wolves, cats, and human beings (Yang et al., Citation2001; da Silva et al., Citation2009; Chen et al., Citation2011). Some Sarcocystis species would cause a lot of severe symptoms in intermediate hosts, like hemorrhagic diathesis and encephalitis (Kolenda et al., Citation2014), while acute infections of pregnant females with Sarcocystis spp. would bring about fetal death, abortion, and premature birth (Dubey et al., Citation1989). Thus Sarcocystosis would cause unedible meat, and vast economic loss further (Xiang et al., Citation2009).
Chinese buffaloes, as the typical ones of swamp buffaloes, are widely distributed in rice growing areas in south of the Huaihe river, and are the intermediate hosts of many Sarcocystis species, including S. fusiformis, S. levinei, S. sinensis, S. cruzi, and S. hominis (Yang et al., Citation2001; Li et al., Citation2002; Yue et al., Citation2013). Among those species, S. fusiformis’ cyst is distinctive from others in its morphology with long spindle shape and length about 5–35 mm (Gjerde et al., Citation2015; Hu et al., Citation2016). S. fusiformis has been distinguished from water buffaloes in Egpyt with 18S rRNA, 28S rRNA, cox1, and internal transcribed spacer 1 (ITS1) gene sequences, yet in Vietnam, Malaysia, Iran, and China with 18S rRNA only (Yang et al., Citation2001; Li et al., Citation2002; Jehle et al., Citation2009; Xiang et al., Citation2009; Oryan et al. Citation2011; Latif et al., Citation2013; Gjerde et al., Citation2015; Hu et al., Citation2016) .
18S rRNA, 28S rRNA, ITS, and cox1 sequences were reported to be effective molecular markers for the identification of Sarcocystis in many countries (Dahlgren & Gjerde, Citation2008; Jehle et al., Citation2009; Oryan et al. Citation2011; Latif et al., Citation2013; Kolenda et al., Citation2014; Gjerde et al., Citation2015; Gjerde & Josefsen, Citation2015). In 2001, 18S rRNA gene sequence was employed to identify S. fusiformis from Chinese buffaloes in Yunnan province, southwest China (Yang et al., Citation2001; Li et al., Citation2002). However, it’s been reported that cox1 sequence is considered to be a better marker than 18S rRNA, as cox1 genes could discriminate closely related Sarcocystis species in 18S rRNA analysis (Gjerde, Citation2014). This study would supply morphological observation with molecular data obtained by sequencing 18S rRNA and cox1 genes from sarcocyst samples found in Guizhou province.
Materials and methods
Sample source
In this study, sarcocysts were isolated from Chinese buffaloes’ muscles in Guanling Buyei and Miao Autonomous County, Guizhou province, southwest of China (East longitude: 105°35′10″–106°0′50″, North latitude: 25°25′19″–26°10′32″), on April, 2016. Cysts were washed three times in PBS and examined through morphological characteristics, then were stored at −20 °C in Laboratory of Parasitology of Northwest A&F University, China, until DNA isolation.
DNA isolation and amplification of 18S rRNA and cox1 genes
After cyst isolation and morphology identification, the extraction of ribosomal RNA and mitochondrial DNA was carried out immediately, with TIANamp Genomic DNA Kit (Tiangen Biotech (Beijing) Co., Ltd, PR China). Isolation procedure was performed according to the manufacturer’s protocol.
The 18S rRNA and cox1 genes were all amplified by polymerase chain reaction (PCR) with primers of 18S rRNA gene (Sense: 5′-TCAGGGAGGTAGTGACAAGA-3′; Anti-sense: 5′-ATGTCTGG ACCTGGTGAGTT-3′) and cox1 gene (Sense: 5′-CTTTAGCGTTGTTGGTAC-3′; Anti-sense: 5′-CCCGTAGGAATGGCAAT-3′). The primers used for 18S rRNA were designed on the basis of published sequences (JQ713824, KT901102, KT901146, KT901160, and KT901169), for cox1 genes (KT901095, KU247899, KT900999, KT901289, KC209696, KC209683, and KC209711), then synthesized at Sangon Biotech Co., Ltd, Shanghai, China. PCR was run with the standard procedure, except set denaturation temperature as 94 °C. The PCR products were analyzed by electrophoresis in 1.5% agarose gel and stained with ethidium bromide. The nucleotide sequences of the 18S rRNA and cox1 genes were determined by the same company mentioned above.
Sequencing and phylogenetic analyses
Successfully sequenced nucleotide sequences were submitted to Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi), and identified by comparing with similar sequences from GenBank database. ClustalX 1.83 and on-line Castresana Lab (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) were used to align the 18S rRNA and cox1 gene sequences in default settings. MEGA 6.06 was used to generate Neighbor-Joining (NJ) phylograms of 18S rRNA and cox1 gene sequences.
Results and discussion
One kind of sarcocysts with long spindle shape was discovered in the muscle samples with different length, longer than 11 mm, shorter than 32 mm, and paralleled with the muscle fibers. The sarcocyst samples are relatively long among Sarcocystis species, with 11–32 mm in length, and long spindle shapes, which are the distinct features of S. fusiformis. S. fusiformis has a prominent size in length and a highly branched cyst wall, while other species (S. levinei, S. sinensis, S. cruzi, and S. hominis) are all smaller with length no longer than 6 mm (Dubey et al. Citation2014).
According to the phylogeny trees, it is obvious that the sarcocysts found in Guizhou province are identical with S. fusiformis sequences. However, this study failed to found S. levinei, S. sinensis, S. cruzi, and S. hominis which were discovered by others in Chinese buffaloes before (Yang et al. Citation2001; Li et al. Citation2002; Hu et al. Citation2016). S. levinei, S. fusiformis, and S. sinensis are common parasites that have been found in China buffaloes, while S. cruzi and S. hominis, mostly found in cattle, are sparsely reported in Bubalus (Hu et al. Citation2016). If the latter two species could parasitize in water buffaloes remains a question, which should be supplemented in further investigation.
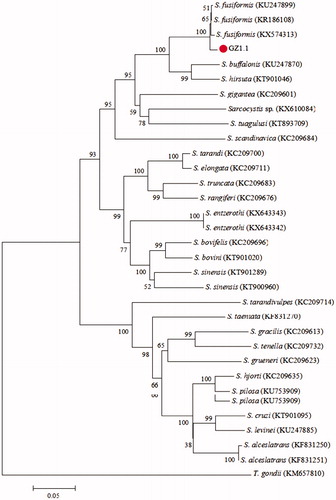
The samples of genomic DNA from sarcocysts were used as template to amplify 18S rRNA gene, which turned out to be 1131 bp long. The obtained sequence was aligned with published sequences of 18S rRNA gene, such as S. fusiformis (KR186119, AF176927, U03071, and KR186116), S. hirsuta (KC209741), and S. silva (KX643339) with an aid of ClustalX 1.83 software. The phylogenetic relationship was established by constructing phylogeny trees obtained with the NJ method, set Bootstrap method as test and Kimura 2-parameter model. The phylogeny tree based on 18S rRNA gene sequences is shown in . The amplifing sequence (named GZ1) was in the same branch with the sequences of S. fusiformis’ 18S rRNA gene. This study used genetic DNA isolated from the specimens as a template to amplify the sequences of cox1 sequence which turned out to be 882 bp long. Like 18S rRNA gene, the amplified cox1 sequence was aligned by ClustalX 1.83 software with the published sequences of cox1 gene like S. sinensis (KT900960, KT901289), S. taeniata (KF831270), and S. bovifelis (KC209696). The phylogeny tree based on cox1 gene sequences is shown in . The cox1 sequence (named GZ1.1) was in the same branch with other sequences of S. fusiformis.
Figure 1. Phylogenetic tree based on partial 18S rRNA sequences of selected Sarcocystidae members and using the NJ method. All sequences’ GenBank accession numbers are given in the brackets after the taxon names. The new sequence of S. fusiformis is indicated by a “•” before the name “GZ1”..
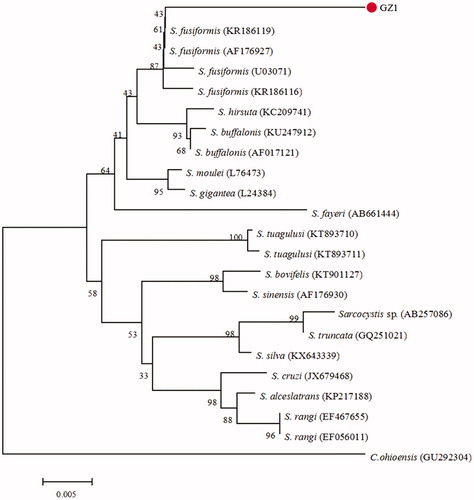
Figure 2. Phylogenetic tree based on partial cox1 sequences of selected Sarcocystidae members and using the NJ method. All sequences’ GenBank accession numbers are given in the brackets after the taxon names. The new sequence of S. fusiformis is indicated by a “•” before the name“GZ1.1”.
There are many researches using molecular methods to identify the sarcocysts in Bubalus in many countries and regions. The most common sequences used to differentiate Sarcocystis species are 18S rRNA, 28S rRNA, cox1 and ITS sequences (Yang et al. Citation2001; Jehle et al. Citation2009; Oryan et al. Citation2011; Latif et al. Citation2013; Gjerde et al. Citation2015). Among them, 18S rRNA and 28S rRNA are highly conservative, and often chosen to be genetic markers to distinguish closely related species, while ITS’s evolvement is relatively rapid, which makes ITS less used than others (Kolenda et al. Citation2014). Moreover, the cox1 gene sequence is affirmed to be more precise than 18S rRNA gene sequence in the phylogenetic discrimination among different species (Dubey et al. Citation1989). Last decades, molecular tests of 18S rRNA were used to reveal the existence of Sarcocystis in many animals, including buffaloes, in many Asian countries like China, Vietnam, India and Iran (Yang et al. Citation2001; Mullaney et al. Citation2005; Jehle et al. Citation2009; Oryan et al. Citation2011; Gjerde Citation2013; Latif et al. Citation2013; Yan et al. Citation2013; Gjerde & Johan Citation2014). In Yunnan China, 28S rRNA and 18S rRNA gene sequence were used to identify the sarcocysts in goats, while in buffaloes, only 18S rRNA was used (Yang et al. Citation2001; Li et al. Citation2002; Hu et al. Citation2016). But in Africa, another continent with serious Sarcocystis prevalence, 18S rRNA, 28S rRNA, cox1 and ITS gene sequences all have been used to discriminate sarcocysts (Gjerde et al. Citation2015). In this study, besides 18S rRNA sequence, cox1 sequence has been employed, as a complement, to examine S. fusiformis in Chinese buffaloes from Guizhou province.
From the phylogenetic trees based on 18S rRNA and cox1 genes, there are high similarity degree between the sample sequence and the sequences of S. fusiformis which were downloaded from NCBI-Nucleotide (https://www.ncbi.nlm.nih.gov/nucleotide/). In , four published sequences of S. fusiformis were used and it turns out the sequences from Egypt (KR186119) and Yunnan China (AF176927) have the closest relationships with GZ1 sequence, while the sequence from Sweden (U03071) shows a little far. This result implied that the 18S rRNA analysis could differentiate the sequences of S. fusiformis from Egypt and Guizhou China, but was not able to distinguish the sequences of S. fusiformis from Egypt, Guizhou China, and Sweden genetically. In , three published sequences of S. fusiformis from Egypt (KU24899 and KR186108) and India (KX574313) were used, and it shows that those sequences and GZ1.1 are in one cluster. This result indicated that the cox1 gene sequence could not discriminate the sequences of S. fusiformis from Egypt, India, and Guizhou China. There is another influencing factor that the samples collected are not enough which can be replenished in further study.
In conclusion, S. fusiformis is identified as one common kind of protozoa of Chinese buffaloes from Guizhou province in this study. And it is also affirmed that 18S rRNA and cox1 genes are effective to differentiate Sarcocystis species from Chinese buffaloes in molecular characterization. This is the first report on molecular analysis using cox1 gene sequence as a genetic marker to affirm the existence of S. fusiformis in Chinese buffaloes in China. The findings in the present study will play an important role in enriching the molecular database of Sarcocystis species worldwide and the diagnosis of sarcocystosis in China.
Acknowledgements
Sincerely thanks people in the investigated regions who assisted in collecting buffalo muscle samples, and others participating in this work.
Disclosure statement
The authors declare that they have no conflicts of interests. The authors alone are responsible for doing the research and writing the paper.
Additional information
Funding
References
- Chen XW, Zuo YX, Rosenthal BM, He YS, Cui LW, Yang ZQ. 2011. Sarcocystis sinensis is an ultrastructurally distinct parasite of water buffalo that can cause foodborne illness but cannot complete its life-cycle in human beings. Vet Parasitol. 178:35–39.
- Dahlgren SS, Gjerde B. 2008. Sarcocystis in moose (Alces alces): molecular identification and phylogeny of six Sarcocystis species in moose, and a morphological description of three new Species. Parasitol Res. 103:93–110.
- Dubey JP, Fayer R, Rosenthal BM, Calero-Bernal R, Uggla A. 2014. Identity of Sarcocystis species of the water buffalo (Bubalus bubalis) and cattle (Bos taurus) and the suppression of Sarcocystis sinensis as a nomen nudum. Vet Parasitol. 205:1–6.
- Dubey JP, Speer CA, Fayer R. 1989. Sarcocystosis of animals and man. Boca Raton (FL): C.A.C. Press Inc.
- Gjerde B. 2013. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int J Parasitol. 43:579–591.
- Gjerde B. 2014. Sarcocystis species in red deer revisited: with a re-description of two known species as Sarcocystis elongata n. sp. and Sarcocystis truncata n. sp. based on mitochondrial cox1 sequences. Parasitol. 141:441–452.
- Gjerde B, Hilali M, Mawgood SA. 2015. Molecular characterisation of three regions of the nuclear ribosomal DNA unit and the mitochondrial cox1 gene of Sarcocystis fusiformis from water buffaloes (Bubalus bubalis) in Egypt. Parasitol Res. 114:3401–3413.
- Gjerde B, Johan S. 2014. Muscular sarcocystosis in two arctic foxes (Vulpes lagopus) due to Sarcocystis arctica n. sp.: sarcocyst morphology, molecular characteristics and phylogeny. Parasitol Res. 113:811–821.
- Gjerde B, Josefsen TD. 2015. Molecular characterisation of Sarcocystis lutrae n. sp. and Toxoplasma gondii from the musculature of two Eurasian otters (Lutra lutra) in Norway. Parasitol Res. 114:873–886.
- Hu JJ, Liu TT, Liu Q, Esch GW, Chen JQ, Huang S, Wen T. 2016. Prevalence, morphology, and molecular characteristics of Sarcocystis spp. in domestic goats (Capra hircus) from Kunming, China. Parasitol Res. 115:3973–3981.
- Hu PF, Jiang WJ, Qu ZW, feng XF, Qian DX, Wang KG. 2016. Pathological diagnosis of Sarcocystosis in buffalo. Heilongjiang Anim Sci Vet Med. 293:93–94.
- Jehle C, Dinkel A, Sander A, Morent M, Romig T, Luc PV, De TV, Thai VV, Mackenstedt U. 2009. Diagnosis of Sarcocystis spp. in cattle (Bos taurus) and water buffalo (Bubalus bubalis) in Northern Vietnam. Vet Parasitol. 166:314–320.
- Kolenda R, Ugorski M, Bednarski M. 2014. Molecular characterization of Sarcocystis species from Polish roe deer based on ssu rRNA and cox1 sequence analysis. Parasitol Res. 113:3029–3039.
- Latif B, Vellayan S, Heo CC, Kannan Kutty M, Omar E, Abdullah S, Tappe D. 2013. High prevalence of muscular sarcocystosis in cattle and water buffaloes from Selangor, Malaysia. Trop Biomed. 30:699–705.
- Li QQ, Yang ZQ, Zuo YX, Attwood SW, Chen XW, Zhang YP. 2002. A PCR-Based RFLP analysis of Sarcocystis cruzi (Protozoa: Sarcocystidae) in Yunnan Province, PR China, reveals the Water Buffalo (Bubalus bubalis) as a natural intermediate Host. J Parasitol. 88:1259–1261.
- Lindsay DS, Verma SK, Scott D, Dubey JP, Dohlen ARV. 2017. Isolation, molecular characterization, and in vitro schizogonic development of Sarcocystis sp. ex Accipiter cooperii from a naturally infected Cooper's hawk (Accipiter cooperii). Parasitol Int. 66:106–111.
- Mullaney T, Murphy AJ, Kiupel M, Bell JA, Rossano MG, Mansfield LS. 2005. Evidence to support horses as natural intermediate hosts for Sarcocystis neurona. Vet Parasitol. 133:27–36.
- Oryan A, Sharifiyazdi H, Khordadmehr M, Larki S. 2011. Characterization of Sarcocystis fusiformis based on sequencing and PCR-RFLP in water buffalo (Bubalus bubalis) in Iran. Parasitol Res. 109:1563–1570.
- da Silva RC, Su CL, Langoni H. 2009. First identification of Sarcocystis tenella (Railliet, 1886) Moulé, 1886 (Protozoa: Apicomplexa) by PCR in naturally infected sheep from Brazil. Vet Parasitol. 165:332–336.
- Xiang Z, Chen XW, Yang LJ, He YS, Jiang RS, Rosenthal BM, Luan PT, Attwood SW, Zuo YX, Zhang YP, Yang ZQ. 2009. Non-invasive methods for identifying oocysts of Sarcocystis spp. from definitive hosts. Parasitol Int. 58:293–296.
- Yan WC, Qian WF, Li XJ, Wang TQ, Ding K, Huang TF. 2013. Morphological and molecular characterization of Sarcocystis miescheriana from pigs in the central region of China. Parasitol Res. 112:975–980.
- Yang ZQ, Zuo YX, Yao YG, Chen XW, Yang GC, Zhang YP. 2001. Analysis of the 18S rRNA genes of Sarcocystis species suggests that the morphologically similar organisms from cattle and water buffalo should be considered the same species. Mol Biochem Parasitol. 115:283–288.
- Yue XP, Li R, Xie WM, Xu P, Chang TC, Liu L, Cheng F, Zhang RF, Lan XY, Chen H, Lei CZ. 2013. Phylogeography and domestication of Chinese swamp buffalo. Plos One. 8:e56552.
