Abstract
Coix lacryma-jobi is a cereal and medicinal crop belonging to the Poaceae family. This study characterized complete chloroplast genome sequence of a Korean cultivar Johyun of C. lacryma-jobi var. ma-yuen through the de novo hybrid assembly with Illumina and PacBio genomic reads. The chloroplast genome is 140,863 bp long and composed of large single copy (82,827 bp), small single copy (12,522 bp), and a pair of inverted repeats (each 22,757 bp). A total of 123 genes including 87 protein-coding genes, 32 tRNA genes, and four rRNA genes were predicted in the genome. Phylogenetic analysis confirmed a close relationship of C. lacryma-jobi with species in the Panicoideae subfamily of the Poaceae family.
Coix lacryma-jobi is an annual herb, called also as adlay or Job’s tears, belonging to the Panicoideae subfamily of the Poaceae family and cultivated in tropical and subtropical regions of the world. C. lacryma-jobi includes many variants such as C. lacryma-jobi var. ma-yuen with soft shell (Chen et al. Citation2006). In Korea, C. lacryma-jobi var. ma-yuen is called as Yulmu and has been cultivated as a cereal and medicinal crop (Park and Lee Citation1999; Kim et al. Citation2000; Lee et al. Citation2002; RDA Citation2013). Several genomic resources have been reported for C. lacryma-jobi, however, those are insufficient for breeding and molecular study of this species, compared to other Panicoideae species such as maize and sorghum whose genomes have been sequenced (Jiao et al. Citation2017; McCormick et al. Citation2018). On this account, this study characterized the complete chloroplast genome of C. lacryma-jobi var. ma-yuen.
A Korean cultivar Johyun of C. lacryma-jobi var. ma-yuen was bred by Gyeonggido Agricultural Research and Extension Services (http://nongup.gg.go.kr/, cultivar no. SJ9203) in Korea (Jang et al. Citation2005) and registered in Korea Seed and Variety Service (https://www.seed.go.kr) under registration no. 05-0020-4. Genomic DNA was extracted from leaves of the cultivar and sequenced using the Illumina HiSeq2500 and PacBio RSII platforms. High quality reads including Illumina paired-end reads of about 5.9 Gb and PacBio reads of about 9.9 Gb were de novo hybrid assembled using SPAdes (ver. 3.9.0, http://cab.spbu.ru/software/spades/) and then chloroplast contigs were selected and sorted by comparison with reported C. lacryma-jobi chloroplast genome sequence (NC_013273, Leseberg and Duvall Citation2009). The selected contigs were merged and gap-filled by a series of read mapping and then chloroplast genome was completed as described by Kim et al. (Citation2015).
The complete chloroplast genome (GenBank Accession no. MH558672) is 140,863 bp long and consists of large single copy (LSC, 82,827 bp), small single copy (SSC, 12,522 bp), and a pair of inverted repeats (IRa and IRb, each 22,757 bp). A total of 123 genes were predicted in the genome, including 87 protein-coding genes, 32 tRNA genes, and four rRNA genes.
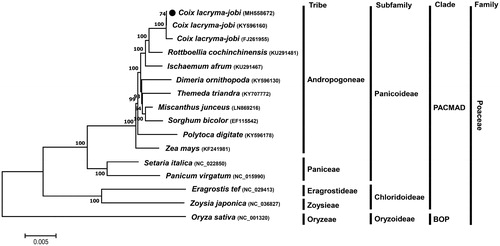
Phylogenetic analysis of C. lacryma-jobi var. ma-yuen cv. Johyun with other taxa was performed using a Maximum Likelihood (ML) method with whole chloroplast genome sequences and revealed that C. lacryma-jobi var. ma-yuen cv. Johyun formed a group with species in the Andropogoneae tribe of the Panicoideae subfamily and was much closed (∼99% identity) to the studied C. lacryma-jobi species (). This phylogenetic relationship was consistent with previous studies (Leseberg and Duvall Citation2009; Teerawatananon et al. Citation2011).
Figure 1. ML phylogenetic tree of chloroplast genomes of C. lacryma-jobi var.ma-yuen cv. Johyun and other Poaceae species. Whole chloroplast genome sequences were multiple-aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate phylogenetic tree by MEGA 7.0 (Kumar et al., Citation2016). The bootstrap support values (>50%) from 1000 replicates are indicated on the nodes. GenBank accession nos. of chloroplast genome sequences used for this tree are indicated within parentheses.
Acknowledgments
We thank Dr. Seung Jae Lee in DNA Link, Inc and Dr. Jihyun Ju in Macrogen, Inc for providing the basic formation of C. lacryma-jobi genome sequencing.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Chen SL, Li DZ, Zhu G, Wu Z, Lu SL, Liu L, Wang ZP, Sun BX, Zhu ZD, Xia N, et al. 2006. Poaceae. In: Wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 22. Beijing (China) and St. Louis (MO): Science Press and Missouri Botanical Garden Press [accessed 2018 June 29] http://www.efloras.org/florataxon.aspx?flora_id =2&taxon_id =10711
- Jang JH, Yi ES, Choi BY, Kim IJ, Park JS, Kim SK, Kim HD. 2005. New variety “Johyun” of Coix lachryma-jobi var. mayuen Stapf with early maturity and short plant height. Korean J Medicinal Crop Sci. 13:122–125.
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. 2017. Improved maize reference genome with single-molecule technologies. Nature. 546:524–527.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655
- Kim HK, Cho DW, Hahm YT. 2000. The effects of Coix Bran on lipid metabolism and glucose challenge in hyperlipidemic and diabetic rats. J Korean Soc Food Sci Nutr. 29:140–146.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee JE, Suh MH, Lee HG, Yang CB. 2002. Characteristics of Job’s tear gruel by various mixing ratio, particle size and soaking time of Job’s tear and rice flour. Korean J Food Cookery Sci. 18:193–199.
- Leseberg CH, Duvall MR. 2009. The complete chloroplast genome of Coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals. J Mol Evol. 69:311–318.
- McCormick RF, Truong SK, Sreedasyam A, Jenkins J, Shu S, Sims D, Kennedy M, Amirebrahimi M, Weers BD, McKinley B, et al. 2018. The Sorghum bicolor reference genome: improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93:338–354.
- Park GS, Lee SJ. 1999. Effect of Job’s tears powder and green tea powder on the characteristics of quality of bread. J Korean Soc Food Sci Nutr. 28:1244–1250.
- Rural Development administration (RDA). 2013. Medicinal plants-Agricultural technique guide 7. Korea: RDA.
- Teerawatananon A, Jacobs SWL, Hodkinson TR. 2011. Phylogenetics of Panicoideae (Poaceae) based on chloroplast and nuclear DNA sequences. Telopea. 13:115–142.
