Abstract
Vitis davidii Foёx is an important wild grape species with extremely high diseases resistance, health promoting properties, and utilization value. Here, the whole-genome high-throughput sequencing data was mined to determine the entire chloroplast genome of this Vitis species. This circular DNA is 161,065 bp in size, counting a pair of inverted repeats (24,802 bp each). The chloroplast genome consists of 125 genes, including 86 protein-coding genes, 31 transfer RNAs, and 8 ribosomal RNAs. A phylogenetic analysis based on the complete chloroplast genome sequences in plants showed that V. davidii formed a different clade from other four congeneric species of the family Vitaceae. This complete chloroplast genome will provide valuable information for future evaluation, conservation, and utilization of V. davidii.
Spine Grape (Vitis davidii Foёx), is one of striking wild germplasms of the East Asian Vitis spp., which is predominantly distributed in South China (Meng et al. Citation2012). This geographical distribution of this species confers their strong tolerance to moisture and heat (Meng et al. Citation2012; Xu et al. Citation2014). Also, this species displays extremely high resistance to Sphaceloma ampelinum de Bary, Coniathyrium diplodilla (Speg.) Sacc.), and Glomerella cigulata (ston.) Spault et Schrenk. Although it is a wild-growing Vitis species, several vines of this species were found to have prominent bisexual flowers. At present, spine grapes were mainly made available as a fresh edible fruit and a small part has also been used for making wines in China. However, it has not yet been effectively developed and fully utilized.
As one of the important genetic marker sources, chloroplast genomes are widely used for phylogenetic analyses, genetic diversity evaluation, and plant molecular identification (Dong et al. Citation2014; Li et al. Citation2016). There are only four chloroplast genomes of the genus Vitis annotated, including V. vinifera (Jansen et al. Citation2006), V. amurensis (Xie et al. Citation2016), V. aestivalis, and V. rotundifolia. In this study, we determined the complete chloroplast genome sequence of V. davidii (GenBank accession no: MG251741), hence adding a new reported chloroplast genome to the genus Vitis. Young and healthy leaves were sampled from V. davidii Foёx in the germplasm nursery (N38°30'43.44", E106°08'19.67") of Ningxia University (Ningxia, China). Genomic DNAs were extracted, quantified and further sequenced on the Illumina Hiseq Xten Platform (Illumina, San Diego, CA). The living material and DNA were stored in Ningxia University, China. The phylogenetic analysis was carried out with the 15 reported plant chloroplast genome sequences using PhyML (Stéphane et al. Citation2005).
The complete chloroplast genome of V. davidii is 161,065 bp long and consists of two inverted repeated regions (IRa and IRb) of 24,802 bp. The chloroplast genome of V. davidii consists of 125 genes, including 86 protein-coding genes, 31 transfer RNAs, and 8 ribosomal RNAs. Intron-exon structure analysis indicated the majority (115 genes, 92%) are genes with no introns, whereas 8 (6.4%) genes contain a single intron and 2 protein-coding genes harbour two introns. The genome organization, and the relative positions of 125 individual genes in chloroplast genome of V. davidii were highly similar to those of the V. vinifera (Number: 84/37/8) (Jaillon et al. Citation2007) and those of the V. amurensis (Number: 88/37/8) (Xie et al. Citation2016) except for some minor differences in numbers of genes.
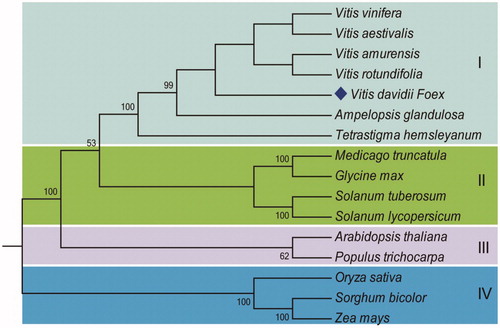
The phylogenetic analysis showed five Vitis species (V. vinifera, V. aestivalis, V. amurensis, V. rotundifolia, and V. davidii), Ampelopsis glaandulosa and a Tetrastigma hemsleyaanum were clustered together with a well-supported cluster, where V. davidii formed a different clade from other four congeneric species of the family Vitaceae (). This suggests that V. davidii is a sister group of the other Vitis species, that split before the divergence among V. vinifera, V. aestivalis, V. amurensis, and V. rotundifolia.
Figure 1. Phylogenetic tree of 16 complete chloroplast genome sequences shows the 4 distinct clusters. GenBank accession numbers: Vitis vinifera (NC_007957), Vitis davidii (MG251741, in this study), Vitis amurensis (KX499471), Vitis aestivalis (KT997470), Vitis rotundifolia (KF976463), Ampelopsis glandulosa (KT831767), Tetrastigma hemsleyanum (KT033563), Sorghum bicolor (NC_008602), Zea mays (NC_001666), Oryza sativa (NC_001320), Medicago truncatula (NC_003119), Glycine max (NC_007942), Solanum tuberosum (NC_008096), Solanum lycopersicum (NC_007898), Arabidopsis thaliana (NC_000932), and Populus trichocarpa (NC_009143).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Dong W, Liu H, Xu C, Zuo Y, Chen Z, Zhou S. 2014. A chloroplast genomic strategy for designing taxon specific DNA mini-barcodes: a case study on ginsengs. BMC Genet. 15:138.
- Jaillon O, Aury J-M, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. 2007. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 449:463.
- Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H. 2006. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 6:32.
- Li B, Li Y, Cai Q, Lin F, Huang P, Zheng Y. 2016. Development of chloroplast genomic resources for Akebia quinata (Lardizabalaceae). Conserv Genet Resour. 8:447–449.
- Meng J, Fang Y, Gao J, Qiao L, Zhang A, Guo Z, Qin M, Huang J, Hu Y, Zhuang X. 2012. Phenolics composition and antioxidant activity of wine produced from spine grape (Vitis davidii Foex) and Cherokee rose (Rosa laevigata Michx.) fruits from South China. J Food Sci. 77:C8.
- Stéphane G, Franck L, Patrice D, Olivier G. 2005. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:557–559.
- Xie H, Jiao J, Fan X, Zhang Y, Jiang J, Sun H, Liu C. 2016. The complete chloroplast genome sequence of Chinese wild grape Vitis amurensis (Vitaceae: Vitis L.). Conserv Genet Resour. 9(1):43–46.
- Xu H, Liu G, Liu G, Yan B, Duan W, Wang L, Li S. 2014. Comparison of investigation methods of heat injury in grapevine (Vitis) and assessment to heat tolerance in different cultivars and species. BMC Plant Biol. 14:156.
