Abstract
The complete chloroplast genome sequence of Codonopsis lanceolata was determined by next generation sequencing. The total length of chloroplast genome of C. lanceolata was 169,447 bp long, including a large single-copy (LSC) region of 85,253 bp, a small single-copy (SSC) region of 8060 bp, and a pair of identical inverted repeat regions (IRs) of 38,067 bp. A total of 110 genes was annotated, resulting in 79 protein-coding genes, 27 tRNA genes, and 4 rRNA genes. The phylogenetic analysis of C. lanceolata with related chloroplast genome sequences in this study provided the taxonomical relationship of C. lanceolata in the genus Campanula.
The genus Codonopsis is composed of ∼42 herbaceous perennial plants belonging to the Campanulaceae family which are native to Central, East and South Asia (Hong et al. Citation2011; He et al. Citation2015). Codonopsis lanceolate is enriched with saponin and has been widely used as medicinal plants effective for bronchitis, cough, spasm, and inflammation (Lee Citation1985; Lee et al. Citation2002). To date, the barcode marker system has not been well established, in spite of the importance of molecular authentication system of medicinal plants, including C. lanceolate. We reported complete chloroplast genome sequence of C. lanceolata which will serve as a source of future barcode marker development, as well as assist further phylogenetic study of genus Codonopsis.
The leaves of C. lanceolata were supplied from Hantaek botanical garden (www.hantaek.co.kr) in Yongin-si, Korea (37° 05′ 40.4” N, 127° 24′ 23.7” E). A genomic library of C. lanceolata was constructed and sequenced using an Illumina HiSeq platform. We obtained a total of 1.6 Gb high-quality paired-end reads and assembled through CLC_assembler (ver. 4.010.83648, CLC QIAGEN) with manual correction by paired-end reads mapping by CLC_mapper (ver. 4.010.83648, CLC QIAGEN) (Kim et al. Citation2015). The genes of the complete chloroplast genome were predicted using Ge-Seq with manual curations of start and stop codons (Tillich et al. Citation2017).
The complete chloroplast genome of C. lanceolata (GenBank accession no. MH018574) has a circular molecular structure of 169,447 bp in length with 38.25% of GC content. Like typical chloroplast genomes, the complete chloroplast genome of C. lanceolata was comprised of four regions: a large single-copy (LSC) region of 85,253 bp, a small single-copy (SSC) region of 8060 bp, and a pair of inverted repeat (IRa and IRb) regions of 38,067 bp. We annotated 110 genes, including 79 protein-coding genes, 27 tRNA genes, and 4 rRNA genes. In addition to gene annotation, 26 simple sequence repeats were identified with the minimum repeat number of 21, 4, and 1 for mono-, di-, and tri- nucleotides, respectively, using MISA tool (http://webblast.ipk-gatersleben.de/misa/) (Beier et al. Citation2017).
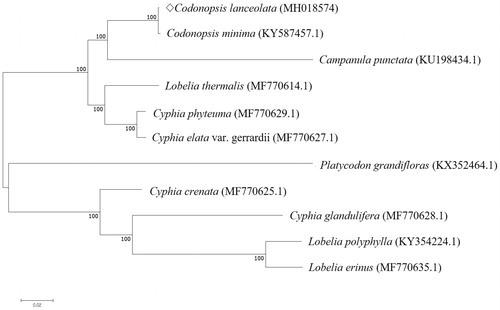
We performed the phylogenetic analysis of C. lanceolata with 10 species of Campanulaceae family using MAFFT (ver. 7.271) by Neighbor-Joining tree analysis of MEGA 7.0 with 1000 bootstraps (Katoh et al. Citation2002; Kumar et al. Citation2016). The phylogenetic analysis revealed genus Codonopsis was closely related to genus Campanula ().
Acknowledgements
We appreciate Hantaek Botanical Garden, Korea, for providing us plant material of Codonopsis lanceolata.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Beier S, Thiel T, Munch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33:2583–2585.
- He JY, Ma N, Zhu S, Komatsu K, Li ZY, Fu WM. 2015. The genus Codonopsis (Campanulaceae): a review of phytochemistry, bioactivity and quality control. J Nat Med. 69:1–21.
- Hong DY, Wu ZY, Raven PH. 2011. Flora of China. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066.
- Kim K, Lee SC, Lee J, Yu Y, Yang K, Choi BS, Koh HJ, Waminal NE, Choi HI, Kim NH, et al. 2015. Complete chloroplast and ribosomal sequences for 30 accessions elucidate evolution of Oryza AA genome species. Sci Rep. 5:15655
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874.
- Lee CB. 1985. Color illustration of Korean plants. Seoul, Korea: Hyangmunsa
- Lee KT, Choi J, Jung WT, Nam JH, Jung HJ, Park HJ. 2002. Structure of a new echinocystic acid bisdesmoside isolated from Codonopsis lanceolata roots and the cytotoxic activity of prosapogenins. J Agric Food Chem. 50:4190–4193.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45:W6–W11.