Abstract
In this work, we present for the first time the mitochondrial genome of a paradoxical frog (Pseudis tocantins). This genome is 15.56 kb, excluding the control region, and is similar in gene content to other hylid mitogenomes. Maximum likelihood analysis, using the mitogenomes of several anurans, indicated P. tocantins as closely related to other hylid species.
The paradoxical frog Pseudis tocantins Caramaschi and Cruz Citation1998 is endemic to Brazilian savannas associated with Araguaia and Tocantins river basins (Frost Citation2018). Interesting features of the paradoxical frogs are related to (1) their extended larval period that results in tadpoles attaining body sizes that are substantially larger than metamorphosed adults (Shaw Citation1802; Garda et al. Citation2010; Santana et al. Citation2016), and (2) the study of sex chromosome differentiation, as P. tocantins has highly heteromorphic sex chromosomes (Busin et al. Citation2008; Gatto et al. Citation2016). Here, we describe the complete mitochondrial genome of P. tocantins.
Total genomic DNA was isolated from the liver of a female P. tocantins collected from the type locality of this species (i.e. Porto Nacional, Tocantins state in Brazil – 10°44′38.6″S 48°26′09.2″W), using a standard phenol:chloroform extraction protocol (Sambrook et al. Citation1989). The remaining tissue is deposited in the tissue collection of the Laboratory of Chromosome Studies of the University of Campinas, Brazil (LabEsC – UNICAMP) under the accession number SMRP 190.15 and the specimen voucher is deposited at the amphibian collection of the Museu de Zoologia Professor Adão José Cardoso at the University of Campinas, Brazil, under the accession number ZUEC 22351. Permission for collecting the specimen was granted by SISBIO (process 45183) and Committee for Ethics in Animal Use of the University of Campinas (CEUA/UNICAMP) (process 3419-1).
A paired-end genomic library was prepared using Nextera DNA Flex Library Prep Kit (Illumina, EUA) and sequenced on an Illumina HiSeq X Ten (Hudson-Alpha Institute for Biotechnology, Alabama, USA). We detected k-mers (k = 31) occurring at high frequencies among genomic reads using RepARK script (Koch et al. Citation2014) and default parameters. These high-frequency k-mers were then assembled by Velvet (Zerbino and Birney Citation2008) using default parameters. Genome annotation was performed using MITOS (Bernt et al. Citation2013). The mitochondrial genome contig was compared with available hylid mitochondrial genomes using BLAST (Altschul et al. Citation1990). The complete mitochondrial genome of P. tocantins, excluding control region, was used in a phylogenetic analysis in comparison with other 13 anurans species. Sequences were aligned using Muscle (Edgar Citation2004) and used for a Maximum likelihood analysis in RAxML v.0.4.1b (Stamakis Citation2014), under the GTR + G evolutionary model.
The assembled mitogenome of P. tocantins (GenBank accession number MH571152) spanned 15.56 kb, with a GC content of 43.16%. This genome is similar to those of other hylids in size, gene content, and arrangement, with an average nucleotide similarity of 70% when compared to six species from Hyla + Dryophytes. There are 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes. These genes are coded in the heavy strand, except for eight tRNA genes (tRNA-Pro, tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser, tRNA-Glu) and one protein-coding gene (ND6).
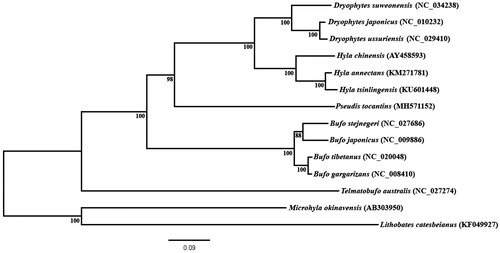
The resulting phylogenetic tree placed P. tocantins as the sister taxon of all extant hylid species analyzed in this paper (). This result agrees with the large-scale phylogenetic inferences published thus far, as they place Pseudis as a member of the Hylidae family (Faivovich et al. Citation2005; Wiens et al. Citation2010; Pyron and Wiens Citation2011; Duellman et al. Citation2016).
Figure 1. Phylogenetic inference obtained using maximum likelihood based on mitochondrial genomes, excluding control region, of the anuran species Dryophytes japonicus, D. ussuriensis, (as Hyla ussuriensis in Sun et al. Citation2017), D. suweonensis (as H. suweonensis in Lee et al. 2017), Hyla chinensis, H. annectans, Hyla tsinlingensis, Pseudis tocantins, Bufo gargarizans, B. tibetanus (as in Wang et al. 2013), stejnegeri, B. japonicus, Telmatobufo australis, Microhyla okinavensis and Lithobates catesbeianus. M. okinavensis and L. catesbeianus were used as outgroups. The phylogenetic inference was constructed under GTR + G evolutionary model. Bootstrap analysis was performed using 1000 pseudoreplicates for node support (numbers at the nodes).
Disclosure statement
No potential conflict of interest was reported by authors.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69:313–319.
- Busin CS, Andrade GV, Bertoldo J, Del Grande ML, Uetanabaro M, Recco-Pimentel SM. 2008. Cytogenetic analysis of four species of Pseudis (Anura, Hylidae), with the description of ZZ/ZW sex chromosomes in P. tocantins. Genetica. 133:119–127.
- Caramaschi U, Cruz CAG. 1998. Notas taxonômicas sobre Pseudis fusca Garman e P. bolbodactyla A. Lutz, com a descrição de uma nova espácie correlata (Anura, Pseudidae). Rev Bras Zool. 15:929–944.
- Duellman WE, Marion AB, Hedges SB. 2016. Phylogenetics, classification, and biogeography of the treefrogs (Amphibia: Anura: Arboranae). Zootaxa. 4104:1–109.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797.
- Faivovich J, Haddad CFB, Garcia PCA, Frost DR, Campbell JA, Wheeler WC. 2005. Systematic review of the frog family Hylidae, with special reference to Hylinae: phylogenetic analysis and taxonomic revision. Bull Am Museum Nat Hist. 294:1–240.
- Frost DR. 2018. Amphibian Species of the World 6.0: An Online Reference. New York: American Museum of Natural History; [accessed 2018 Jul 06]. http://research.amnh.org/vz/herpetology/amphibia/.
- Garda AA, Santana DJ, São-Pedro V. d A. 2010. Taxonomic characterization of paradoxical frogs (Anura, Hylidae, Pseudae): geographic distribution, external morphology, and morphometry. Zootaxa. 2666:1–28.
- Gatto KP, Busin CS, Lourenço LB. 2016. Unraveling the sex chromosome heteromorphism of the paradoxical frog Pseudis tocantins. PLoS One. 11:e0156176.
- Koch P, Platzer M, Downie BR. 2014. RepARK – de novo creation of repeat libraries from whole-genome NGS reads. Nucleic Acids Res. 42:e80.
- Pyron RA, Wiens JJ. 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol Phylogenet Evol. 61:543–583.
- Sambrook J, Fritschi EF, Maniats T. 1989. Molecular cloning: a laboratory manual. 2nd ed. New York (NY): Cold Spring Harbor Laboratory Press.
- Santana DJ, Magalhães F. de M, São-Pedro V. de A, Mângia S, Amado TF, Garda AA. 2016. Calls and tadpoles of the species of Pseudis (Anura, Hylidae, Pseudae). Herpetol J. 26:139–148.
- Shaw G. 1802. General zoology or systematic natural history. Volume III, Part 1. Amphibia. London (UK): Thomas Davison.
- Stamakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30: 1312–1313.
- Sun Q, Xie Y, Zhao W, Liu P. 2017. Sequencing and analysis of the complete mitochondrial genome of Hyla ussuriensis (Anura: Hylidae). Mitochondr DNA part A. 28: 328–329.
- Wang X, Wang Y, Yue B, Zhang X, Liu S. 2017. The complete mitochondrial genome of the Bufo tibetanus (Anura: Bufonidae). Mitochondr DNA. 24: 186–188.
- Wiens JJ, Kuczynski CA, Hua X, Moen DS. 2010. An expanded phylogeny of treefrogs (Hylidae) based on nuclear and mitochondrial sequence data. Mol Phylogenet Evol. 55:871–882.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18:821–829.
