Abstract
The complete mitochondrial DNA genome of the Salangid icefish (Neosalanx taihuensis) was sequenced by the primer walking sequence method. The entire mitochondrial genome of this species is 17,035 bp in length, making it the longest among the reported mitochondrial genomes of Osmeriformes. It contains 13 protein-coding genes, 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and one control region (CR). The gene arrangement, nucleotide composition, and codon usage pattern of the mitochondrial genome are similar to those of other teleosts except for two long tandem repeats in the CR. A 486 bp tandem repeat fragment was identified that comprises 2 copies of 243 bp motif and accounts approximately 35.5% of the CR. The 243 bp tandem repeat motif can be folded into a stem-loop secondary structure. Phylogenetic analysis based on 12 concatenated protein-coding genes of the heavy strand shows the genus Neosalanx diverged most recently and clustered with Protosalanx hyalocranius as a clade.
Neosalanx taihuensis belongs to the genus Neosalanx within the family Salangidae of the order Osmeriformes, which is an economically important fish. This species is endemic to China and inhabits mainly the middle and lower reaches of the Yangtze River, including its tributaries and affiliated lakes (Froese and Pauly Citation2017) . In recent years, because of water pollution, overfishing and habitat destruction, the population of N. taihuensis has undergone the loss of appropriate habitat and the decrease in population size (Li Citation2001; Wang et al. Citation2005). Therefore, the genetic information of N. taihuensis is very urgent to demand.
The fish was obtained from the Taihu Lake (120°19′E, 31°25′N), Jiangsu Province in 2012. After morphological identification, the specimen was deposited in our laboratory (Animal genetics lab, Jianghan University). The fin clips were sampled and then preserved in 95% ethanol. Total genomic DNA was extracted using standard phenol-chloroform methods (Sambrook and Russell Citation2001). Seven sets of primers were designed from the conservative regions based on the alignment of complete mitochondrial genomes available within the family Salangidae.
The complete mitochondrial genome sequence of N. taihuensis was 17,035 bp in length (GenBank Accession number MH348204). The protein-coding genes, rRNA genes, tRNA genes, and one control region (CR) of the mitochondrial genome were annotated using MitoAnnotator (Iwasaki et al. Citation2013). The mitochondrial genome of N. taihuensis consisted of 13 typical vertebrate protein-coding genes, 22 tRNAs, 2 rRNAs, and a CR. The gene order and gene content of the mitochondrial genome of N. taihuensis are similar to those of other teleosts (Broughton et al. Citation2001; Zhong et al. Citation2015). Among the 13 protein-coding genes, ten mitochondrial genes (ND1, ND2, COX1, ATP8, ATP6, COX3, ND3, ND4L, ND5, and ND6) are encoded by the typical TAA or TAG stop codons, while the remaining three mitochondrial genes are encoded by an incomplete stop codon T(aa). The CR is a 1369 bp sequence with two long tandem repeats located between the tRNA-Pro and tRNA-Phe genes, which is the longest among the CR reported of the family Salangidae. The tandem repeat motif is 243 bp long, which can be folded into a stable stem-loop secondary structure.
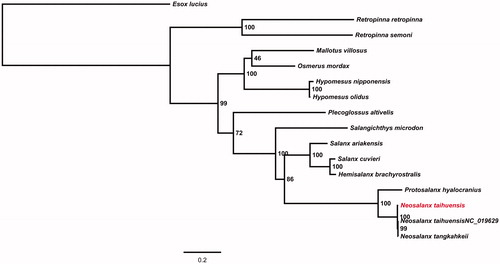
To explore the evolutionary status of N. taihuensis within the order Osmeriformes, a maximum likelihood (ML) tree was constructed using RAxML version 8.1.17 provided by Heidelberg Institute for Theoretical Studies, Heidelberg, Germany (Stamatakis Citation2014). Phylogenetic tree showed that the genus Neosalanx diverged most recently and clustered with Protosalanx hyalocranius as a clade (). In conclusion, the information of N. taihuensi mitochondrial genome reported here may facilitate further investigations of molecular evolution of species in the order Osmeriformes.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Broughton RE, Milam JE, Roe BA. 2001. The complete sequence of the zebrafish (Danio rerio) mitochondrial genome and evolutionary patterns in vertebrate mitochondrial DNA. Genome Res. 11:1958–1967.
- Froese R, Pauly D. 2017. FishBase. World Wide Web electronic publication. http://www.fishbase.org, version (2/2017).
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30:2531–2540.
- Li S. 2001. A study on biodiversity and its conservation of major fishes in the Yangtze River. Shanghai, China: Shanghai Scientific and Technical Publishers.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. New York, NY: Cold Spring Harbor Press.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Wang Z, Lu C, Hu H, Zhou Y, Xu C, Lei G. 2005. Freshwater icefishes (Salangidae) in the Yangtze River basin of China: spatial distribution patterns and environmental determinants. Environ Biol Fish. 73:253–262.
- Zhong L, Wang M, Li D, Tang S, Zhang T, Bian W, Chen X. 2015. Complete mitochondrial genome of Chinese icefish Neosalanx tangkahkeiis (Salmoniformes, Salangidae): comparison reveals Neosalanx taihuensis not a valid name. Mitochondrial DNA Part A, DNA Mapping, Sequencing, and Analysis. 27:3303–3305.