Abstract
Carinotetraodon travancoricus was classified as vulnerable on the IUCN Red List due to habitat loss and overharvesting for the aquarium trade. To gain its molecular information and thus contribute to help in conserving this vulnerable species, we determined the complete mitochondrial DNA of the C. travancoricus. The size of the molecule is 16,542 nucleotides, containing 13 protein-coding genes, 2 rRNA genes, 22 tRNA genes, a putative control region, and 1 origin of replication on the light-strand. The overall base composition includes C(28.8%), A(28.5%), T(27.1%), and G(15.6%). Moreover, the 13 PCGs encode 3800 amino acids in total. The result of the phylogenetic tree supports C. octofasciatus has a closest relationship with Tetraodon nigroviridis.
Carinotetraodon travancoricus is endemic to the Western Ghats of India, and is found in the coastal areas of Kerala and southern Karnataka (Doi et al. Citation2014). The habitat of this fish is severely modified by damming, indiscriminate de-forestation and subsequent conversion of forest area into agricultural plantations, capturing the species for aquarium trade is also a threat. So, C. travancoricus was classified as vulnerable on the IUCN Red List due to habitat loss and overharvesting for the aquarium trade (Raghavan et al. Citation2013). In this study, we determined and described the complete mitochondrial genome of C.travancoricus and explored the phylogenetic relationship within Tetraodontidae, to gain its molecular information and thus contribute to help in conserving this vulnerable species.
The specimen was collected from Periyar River, India (10°10′36″N; 76°9′46″E) and stored in the laboratory of Zhejiang Ocean University with accession number 20150826WW22. Similar to the typical mitogenome of vertebrates, the mitogenome of C. octofasciatus is a closed double-stranded circular molecule of 16,542 nucleotides (GenBank accession No. MH631014), within the range of other teleost mitogenomes. As in other vertebrate (Miya et al. Citation2001; Du et al. Citation2018; Zhu et al. Citation2018a), it contains 13 PGGs, 2 ribosomal RNA genes (12S rRNA and 16S rRNA), 22 tRNA genes, and 2 main non-coding regions. The overall base composition is 28.5% A, 28.8% C, 27.1% Tand 15.6% G respectively, with a slight AT bias (55.6%). most mitochondrial genes of C. octofasciatus were encoded on the H-strand, with only ND6 and eight tRNA (Gln, Ala, Asn, Cys, Tyr, Ser-UCN, Glu, Pro) genes encoded on the L-strand. Thirteen PCGs genes encode 3800 amino acids in total, all of them use the initiation codon ATG except COI use GTG, which is quite common in vertebrate mtDNA (Zhu et al. Citation2018b). Most of them have TAA and TAG as the stop codon, whereas ND3 ends with TAG, whereas COI and ND6 ended with AGG, and two protein-coding genes (COII, ND4) ended with a single T. The lengths of 12S rRNA located between tRNAPhe and tRNAVal and 16S rRNA located between tRNAVal and tRNALeu were 957 bp and 1737 bp respectively. The origin of light-strand replication is located in a cluster of five tRNA genes (WANCY) as in other vertebrates (Zhu et al. Citation2018c), which has the potential to fold into a stable stem-loop secondary structure, with a stem formed by 12 paired nucleotides and a loop of 11 nucleotides; The CR is determined to be 826 bp, which is located between the tRNA-Pro and tRNA-Phe genes, and three typical domains are observed, including the TAS, central CSB and CSB, which is identical to that in other teleostean mitogenomes (Zhang et al. Citation2013).
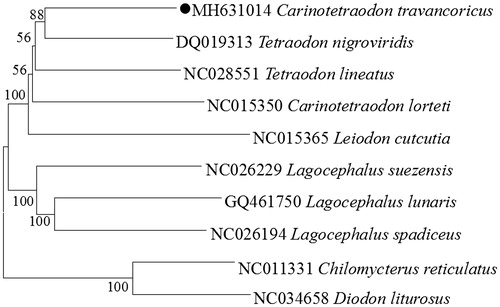
The phylogenetic tree based on the Neighbour-Joining method was constructed to provide relationship within Tetraodontidae. The result of the present study supports C. octofasciatus has a closest relationship with Tetraodon nigroviridis, highly supported by a bootstrap probability of 88% (), which is in accord with the previously reported study (Santini et al. Citation2013).
Figure 1. Neighbor Joining (NJ) tree of 8 Tetraodontidae species based on 12 PCGs. The bootstrap values are based on 1000 re-samplings. The number at each node is the bootstrap probability. The number before the species name is the GenBank accession number. The genome sequence in this study is labeled with a black spot.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Reference
- Doi H, Sakai H, Yamanoue Y, Sonoyama T, Ishibashi T. 2014. Spawning of eight Southeast Asian brackish and freshwater puffers of the genera Tetraodon and Carinotetraodon in captivity. Fisheries Science. 81:291–299.
- Du X, Gong L, Chen W, Liu L, Lü Z. 2018. The complete mitochondrial genome of Epinephelus chlorostigma (Serranidae; Epinephelus) with phylogenetic consideration of Epinephelus. Mitochondrial DNA Part B. 3:209–210.
- Miya M, Kawaguchi A, Nishida M. 2001. Mitogenomic exploration of higher teleostean phylogenies: a case study for moderate-scale evolutionary genomics with 38 newly determined complete mitochondrial DNA sequences. Mol Biol Evol. 18:1993–2009.
- Raghavan R, Dahanukar N, Tlusty MF, Rhyne AL, Kumar KK, Molur S, Rosser AM. 2013. Uncovering an obscure trade: threatened freshwater fishes and the aquarium pet markets. Biol Conserv. 164:158–169.
- Santini F, Sorenson L, Alfaro ME. 2013. A new phylogeny of tetraodontiform fishes (Tetraodontiformes, Acanthomorpha) based on 22 loci. Mol Phylogenet Evol. 69:177–187.
- Zhang H, Zhang Y, Zhang X, Song N, Gao T. 2013. Special structure of mitochondrial DNA control region and phylogenetic relationship among individuals of the black rockfish, Sebastes schlegelii. Mitochondrial DNA. 24:151–157.
- Zhu K, Gong L, Jiang L, Liu L, Lü Z, Liu B-j. 2018a. Phylogenetic analysis of the complete mitochondrial genome of Anguilla japonica (Anguilliformes, Anguillidae). Mitochondrial DNA Part B. 3:536–537.
- Zhu K, Gong L, Lü Z, Liu L, Jiang L, Liu B. 2018b. The complete mitochondrial genome of Chaetodon octofasciatus (Perciformes: Chaetodontidae) and phylogenetic studies of Percoidea. Mitochondrial DNA Part B. 3:531–532.
- Zhu K, Lü Z, Liu L, Gong L, Liu B. 2018c. The complete mitochondrial genome of Trachidermus fasciatus (Scorpaeniformes: Cottidae) and phylogenetic studies of Cottidae. Mitochondrial DNA Part B. 3:301–302.
