Abstract
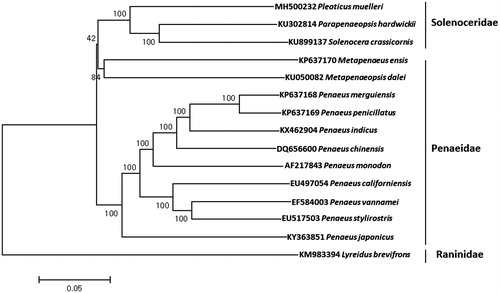
The complete mitochondrial genome of Argentine red shrimp, Pleoticus muelleri (Bate, 1888) was determined using the MiSeq platform. Its mitogenome (16,189 bp) encoded the canonical 13 protein-coding genes, 22 tRNA genes, two rRNA genes. Start codons of all protein-coding genes were ATN except for ATP8, while incomplete stop codons (T–) were identified in six genes including COXI, COXII, COXIII, NAD3, NAD5, and NAD6. The mitogenome of P. muelleri showed highest sequence identity with Parapenaeopsis hardwickii (82%) and Solenocera crassicornis (81%). Phylogenetic analysis showed three shrimps in Solenoceridae including P. muelleri were grouped together from those in Penaeidae, which suggests taxonomic reexamination for those in those family.
The Argentine red shrimp, Pleoticus muelleri is one of the most commercially imported shrimps in Argentina (Bertuche et al. Citation2005). This shrimp was distributed mainly in the Southwestern Atlantic Ocean from the coast of Santa Cruz, Argentina north to Rio de Janeiro, Brazil (Stamatopoulos Citation1993). Its bottom trawl fishery is now recommended to be prohibited to reduce the devastating impacts on benthic ecosystem with high bycatch as well as overfishing. Although this species is now exported in many countries, genetic study of P. muelleri has not been conducted (Batista et al. Citation2011; Azevedo and Barbosa Citation2016). As the first step, the full mitochondrial genome of P. muelleri was determined for the first time among the genus Pleoticus.
The frozen P. muelleri was purchased from an Argentina importer in July 2016 as the part of project to identify the origin of fisheries products in Korea. Its identification was confirmed by both morphological analysis and its 100% match of COI sequence to database (GenBank number MF490134). Examined specimen was stored at National Institute of Fisheries Science (NIFS). The full mitochondrial genome sequence was obtained by assembling two large PCR fragments (COI ∼ ND1: 10,386 bp, ND1 ∼ COI: 7377 bp) amplified by two primer sets designed from the multiple alignments of 13 decapod mitogenome sequences. The PCR products were pooled together in equal concentration and fragmented into 350 bp in length by Covaris® M220 (Covaris Inc., Woburn, MA). A library was constructed by TruSeq® RNA library preparation kit V2 (Illumina, San Diego, CA) and MiSeq Sequencer (Illumina, San Diego, CA) was used for NGS sequencing. Raw reads were trimmed and reads with low quality (QV < 20) were truncated at CLC Genomic Workbench v8.0 (CLC Bio, Cambridge, MA). Mitogenome was reconstructed with Geneious® 11.0.2 (Kearse et al. Citation2012). The mitogenome of Parapenaeopsis hardwickii (KU302814) was employed as a reference sequence (Mao et al. Citation2016). The phylogenetic tree was constructed by MEGA 7 software (Kumar et al. Citation2016) with Minimum Evolution (ME) algorithm.
The mitochondrial genome of P. muelleri was 16,189 bp in length (GenBank Number: MH500232), which encodes 13 proteins, 22 tRNAs, and two ribosomal RNAs (12S and 16S ribosomal RNA) and a putative control region (D-loop). The proportions of A + T (68.22%) was much higher than G + C (31.78%). Total 10 genes were located at L strand and remaining 27 genes were at H strand. Overlapping protein-coding genes were detected between ATP8 ∼ ATP6 (6 bp) and ND4 ∼ ND4L (6 bp). Start codons of all protein-coding genes were ATN except for ATP8, while incomplete stop codons (T–) were identified in six genes including COXI, COXII, COXIII, NAD3, NAD5, and NAD6. Twenty-two tRNAs were predicted to be a typical folded clover-leaf secondary structure (Laslett and Canbäck Citation2008). As the result of phylogenetic analysis, P. muelleri was clustered together with Parapenaeopsis hardwickii (82%) and Solenocera crassicornis (81%) from those in family Penaeidae, which suggests further taxonomic reexamination of shrimps in superfamily Penaeoidea ().
Figure 1. Phylogenetic tree of Pleoticus muelleri with other marine shrimps. The phylogenetic tree was constructed with the complete mitochondrial genome sequence of Pleoticus muelleri by using MEGA 7 software with Minimum Evolution (ME) algorithm with 1000 bootstrap replications. The mitochondrial genome sequences were obtained from the GenBank database.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Azevedo V, Barbosa C. 2016. Argentine Red Shrimp (Pleoticus muelleri) Report.
- Batista AC, Simões SM, Lopes M, Costa RCd. 2011. Ecological distribution of the shrimp Pleoticus muelleri (Bate, 1888) and Artemesia longinaris Bate, 1888 (Decapoda, Penaeoidea) in the southeastern Brazilian littoral. Nauplius. 19:135–143.
- Bertuche D, De La Garza J, Fernández M, Moriondo P, Piñero R, Roux A. 2005. Estudio de las potenciales afectaciones de las distintas actividades económico-productivas realizadas en la zona costera patagónica, en especial en el Golfo San Jorge, sobre las especies bentónicas, en relación a la evolución de aquellas definidas como indicadoras. Consultora Serman & Asociados SA, Informe Proyecto Langostino, INIDEP.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28:1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24:172–175.
- Mao Z-C, Liu P, Duan Y-F, Li J, Chen P, Meng X-L. 2016. Sequencing of complete mitochondrial genome of sword prawn Parapenaeopsis hardwickii (Miers) (Crustacea, Decapoda, Penaeidae). Mitochondrial DNA B. 1:259–260.
- Stamatopoulos C. 1993. Trends in catches and landings. 1. Atlantic fisheries: 1970–1991. Rome: FAO.
